@zsh-o
2018-08-06T14:32:56.000000Z
字数 4094
阅读 1521
9 - EM - GMM - 代码
《统计学习方法》
该部分以代码为主,EM以前有写过,原理部分请参照Expectation Maximisation (EM)
%matplotlib inlineimport numpy as npfrom matplotlib import pyplot as pltepsilon = 1e-8
EM推导完之后的形式还是比较简单的,现以GMM为例
E-step求出现有观测数据下每个观测数据隐变量的最优的分布函数
M-step在上面求得每一个观测数据的的情况下,求当前最优的参数
最后不断迭代上述过程即可
from sklearn.datasets import make_blobs
## 生成数据X, Y = make_blobs(n_samples=1000,n_features=2, centers=3)plt.scatter(X[:, 0], X[:, 1], marker='.', c=Y, s=25, cmap='Dark2')
<matplotlib.collections.PathCollection at 0x1d1a6eda630>
class GMM(object):def __init__(self, K, max_iter=500, eps=1e-5):self.K = Kself.max_iter = max_iterself.eps = epsdef gaussian(self, X, mu, sigma):return 1. / (np.power(2*np.pi, self.D/2.)* np.sqrt(np.linalg.det(sigma))) * np.exp(-0.5 * np.sum(np.dot(X - mu, np.linalg.pinv(sigma))* (X - mu), axis=1))def E_step(self):self.Q = self.pdf * self.Piself.Q = self.Q / np.sum(self.Q, axis=1, keepdims=True)def M_step(self):for k in range(self.K):t = np.sum(self.Q[:, k])self.Pi[k] = t / self.Nself.Mu[k] = np.sum(self.X * self.Q[:, k][:, np.newaxis], axis=0) / tself.Sigma[k] = np.dot((self.Q[:, k][:, np.newaxis] * (self.X - self.Mu[k])).T, self.X - self.Mu[k]) / tdef log_likehood(self):return np.sum(np.log(np.sum(self.pdf * self.Pi, axis=1)))def fit(self, X):self.X = Xself.N, self.D = X.shapeself.Q = np.zeros((self.N, self.K))self.Pi = np.zeros(self.K) + 1./self.Kself.Mu = np.random.permutation(self.X)[:self.K] ## 随机选择K个当作初始均值self.Sigma = np.concatenate([np.cov(self.X.T)[np.newaxis,:] for _ in range(self.K)])self.pdf = np.zeros((self.N, self.K))old_loglikehood = np.inffor _iter in range(self.max_iter):for k in range(self.K):self.pdf[:, k] = self.gaussian(self.X, self.Mu[k], self.Sigma[k])self.E_step()self.M_step()new_llh = self.log_likehood()if np.abs(old_loglikehood - new_llh) < self.eps:print('End with Eps at iter %d' % _iter)returnold_loglikehood = new_llhprint('End with max iter %d' % self.max_iter)def p(self, x):if x.ndim < 2:x = x[np.newaxis, :]N = x.shape[0]t = np.zeros((N, self.K))for k in range(self.K):t[:, k] = self.Pi[k] * self.gaussian(x, self.Mu[k], self.Sigma[k])return np.sum(t, axis=1)def predict(self, x):if x.ndim < 2:x = x[np.newaxis, :]N = x.shape[0]t = np.zeros((N, self.K))for k in range(self.K):t[:, k] = self.Pi[k] * self.gaussian(x, self.Mu[k], self.Sigma[k])return np.argmax(t, axis=1), t / np.sum(t, axis=1)
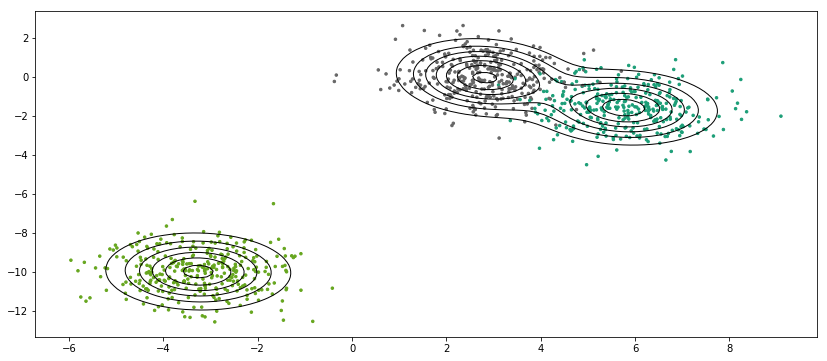
gmm = GMM(K=3)gmm.fit(X)
End with Eps at iter 75
def plot_decision_boundary(model, resolution=100, colors=('b', 'k', 'r'), figsize=(14,6)):plt.figure(figsize=figsize)xrange = np.linspace(model.X[:,0].min(), model.X[:,0].max(), resolution)yrange = np.linspace(model.X[:,1].min(), model.X[:,1].max(), resolution)grid = [[model.p(np.array([xr, yr])) for yr in yrange] for xr in xrange]grid = np.array(grid).reshape(len(xrange), len(yrange))plt.scatter(model.X[:, 0], model.X[:, 1], marker='.', c=Y-1, s=25, cmap='Dark2')plt.contour(xrange, yrange, grid.T, linewidths=(1,),linestyles=('-',), colors=colors[1])
gmm.Y = Y
plot_decision_boundary(gmm)
from sklearn.datasets import make_moonsfrom sklearn.datasets import make_circlesfrom sklearn.preprocessing import StandardScaler
X, Y = make_moons(n_samples=500, shuffle=True, noise=0.1)
gmm = GMM(K=2)gmm.fit(X)
End with Eps at iter 116
gmm.Y = Yplot_decision_boundary(gmm)
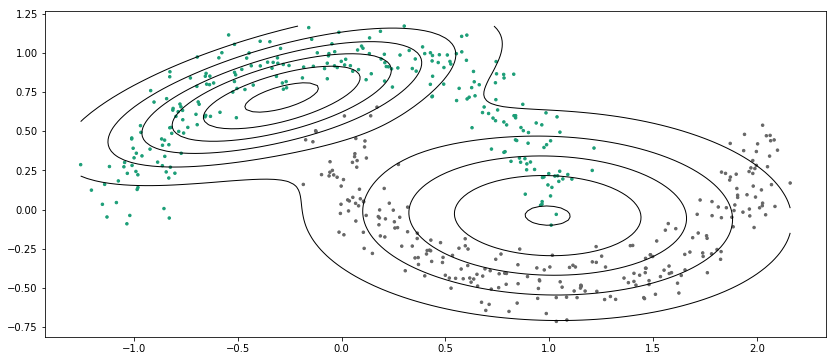
最后说一点,GMM还是有很多地方可以向下做的,例如不用指定聚类数量的无参模型(狄利克雷过程高斯混合模型),另一个是,现有高斯混合模型只能聚类类高斯分布的数据,而其他形状的数据,例如前面SVM节里面的环形和半月型数据就不再适用,但有模型叫结构化的变分自编码,其用多层NN把数据映射到隐状态空间(变分自编码),然后在隐状态空间用高斯混合模型进行聚类,用以发现隐藏在数据中的本质属性,因此可以适用于任意形状的数据
