@EtoDemerzel
2017-11-22T12:30:47.000000Z
字数 6713
阅读 2101
机器学习week8 ex7 review
机器学习 吴恩达
这周学习K-means,并将其运用于图片压缩。
1 K-means clustering
先从二维的点开始,使用K-means进行分类。
1.1 Implement K-means
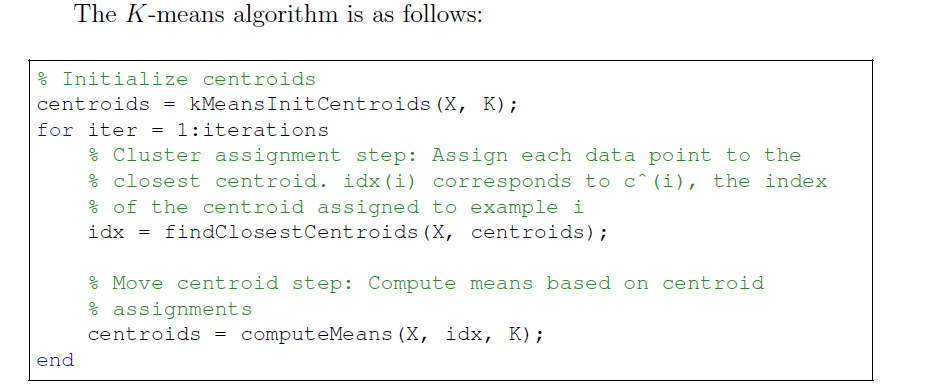
K-means步骤如上,在每次循环中,先对所有点更新分类,再更新每一类的中心坐标。
1.1.1 Finding closest centroids
对每个example,根据公式:

找到距离它最近的centroid,并标记。若有数个距离相同且均为最近,任取一个即可。
代码如下:
function idx = findClosestCentroids(X, centroids)%FINDCLOSESTCENTROIDS computes the centroid memberships for every example% idx = FINDCLOSESTCENTROIDS (X, centroids) returns the closest centroids% in idx for a dataset X where each row is a single example. idx = m x 1% vector of centroid assignments (i.e. each entry in range [1..K])%% Set KK = size(centroids, 1);% You need to return the following variables correctly.idx = zeros(size(X,1), 1);% ====================== YOUR CODE HERE ======================% Instructions: Go over every example, find its closest centroid, and store% the index inside idx at the appropriate location.% Concretely, idx(i) should contain the index of the centroid% closest to example i. Hence, it should be a value in the% range 1..K%% Note: You can use a for-loop over the examples to compute this.%for i = 1:size(X,1)dist = pdist([X(i,:);centroids])(:,1:K);[row, col] = find(dist == min(dist));idx(i) = col(1);end;
1.1.2 Compute centroid means
对每个centroid,根据公式:
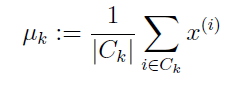
求出该类所有点的平均值(即中心点)进行更新。
代码如下:
function centroids = computeCentroids(X, idx, K)%COMPUTECENTROIDS returns the new centroids by computing the means of the%data points assigned to each centroid.% centroids = COMPUTECENTROIDS(X, idx, K) returns the new centroids by% computing the means of the data points assigned to each centroid. It is% given a dataset X where each row is a single data point, a vector% idx of centroid assignments (i.e. each entry in range [1..K]) for each% example, and K, the number of centroids. You should return a matrix% centroids, where each row of centroids is the mean of the data points% assigned to it.%% Useful variables[m n] = size(X);% You need to return the following variables correctly.centroids = zeros(K, n);% ====================== YOUR CODE HERE ======================% Instructions: Go over every centroid and compute mean of all points that% belong to it. Concretely, the row vector centroids(i, :)% should contain the mean of the data points assigned to% centroid i.%% Note: You can use a for-loop over the centroids to compute this.%for i = 1:Kcentroids(i,:) = mean(X(find(idx == i),:));end;% =============================================================end
1.2 K-means on example dataset
ex7.m中提供了一个例子,其中中 K 已经被手动初始化过了。
% Settings for running K-MeansK = 3;max_iters = 10;% For consistency, here we set centroids to specific values% but in practice you want to generate them automatically, such as by% settings them to be random examples (as can be seen in% kMeansInitCentroids).initial_centroids = [3 3; 6 2; 8 5];
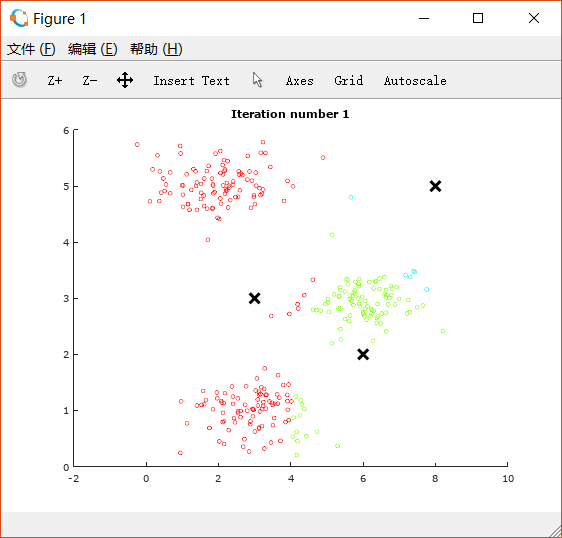
如上,我们要把点分成三类,迭代次数为10次。三类的中心点初始化为.
得到如下图像。(中间的图像略去,只展示开始和完成时的图像)
这是初始图像:
进行10次迭代后的图像:
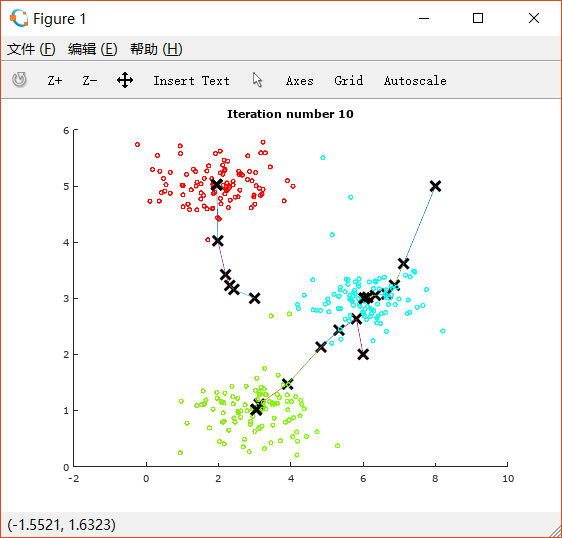
可以看到三堆点被很好地分成了三类。图片上同时也展示了中心点的移动轨迹。
1.3 Random initialization
ex7.m中为了方便检验结果正确性,给定了K的初始化。而实际应用中,我们需要随机初始化。
完成如下代码:
function centroids = kMeansInitCentroids(X, K)%KMEANSINITCENTROIDS This function initializes K centroids that are to be%used in K-Means on the dataset X% centroids = KMEANSINITCENTROIDS(X, K) returns K initial centroids to be% used with the K-Means on the dataset X%% You should return this values correctlycentroids = zeros(K, size(X, 2));% ====================== YOUR CODE HERE ======================% Instructions: You should set centroids to randomly chosen examples from% the dataset X%% Initialize the centroids to be random examples% Randomly reorder the indices of examplesrandidx = randperm(size(X, 1));% Take the first K examples as centroidscentroids = X(randidx(1:K), :);% =============================================================end
这样初始的中心点就是从X中随机选择的K个点。
1.4 Image compression with K-means
用K-means进行图片压缩。
用一张的图片为例,采用RGB,总共需要个bit。
这里我们对他进行压缩,把所有颜色分成16类,以其centroid对应的颜色代替整个一类中的颜色,可以将空间压缩至 个bit。
用题目中提供的例子,效果大概如下:
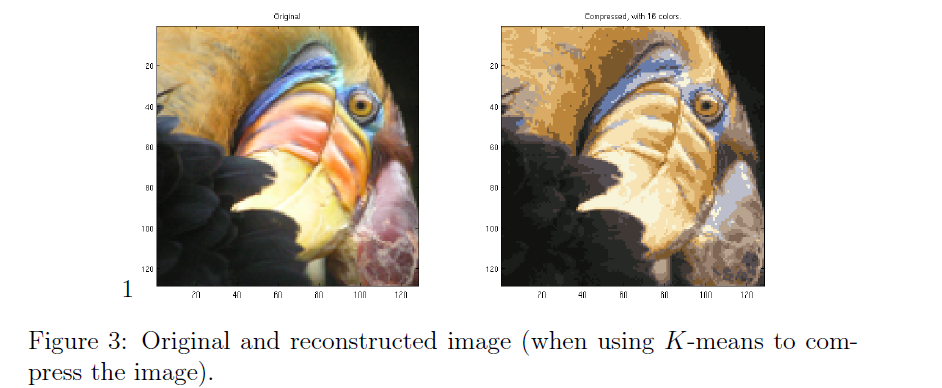
1.5 (Ungraded)Use your own image
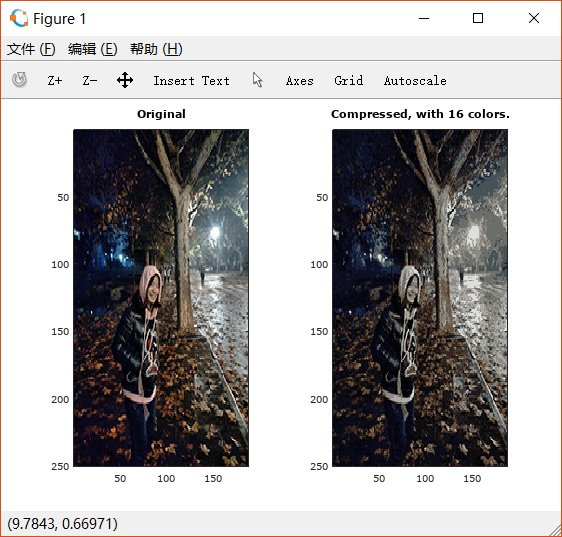
随便找一张本地图片,先用PS调整大小,最好在 以下(否则速度会很慢),运行,效果如下:
2 Principal component analysis
我们使用PCA来减少向量维数。
2.1 Example dataset
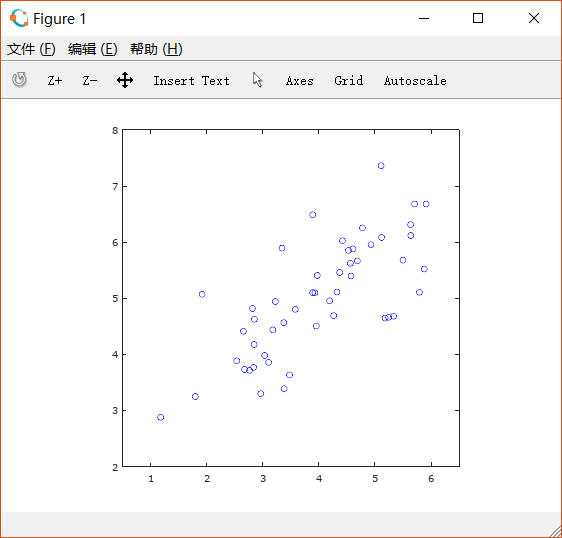
先对例子中的二维向量实现降低到一维。
绘制散点图如下:
2.2 Implementing PCA
首先需要计算数据的协方差矩阵(covariance matrix)。
然后使用 Octave/MATLAB中的SVD函数计算特征向量(eigenvector)。
可以先对数据进行normalization和feature scaling的处理。
协方差矩阵如下计算:
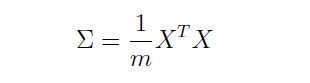
然后用SVD函数求特征向量。
故完成pca.m如下:
function [U, S] = pca(X)%PCA Run principal component analysis on the dataset X% [U, S, X] = pca(X) computes eigenvectors of the covariance matrix of X% Returns the eigenvectors U, the eigenvalues (on diagonal) in S%% Useful values[m, n] = size(X);% You need to return the following variables correctly.U = zeros(n);S = zeros(n);% ====================== YOUR CODE HERE ======================% Instructions: You should first compute the covariance matrix. Then, you% should use the "svd" function to compute the eigenvectors% and eigenvalues of the covariance matrix.%% Note: When computing the covariance matrix, remember to divide by m (the% number of examples).%[U,S,V] = svd(1/m * X' * X);% =========================================================================end
把求出的特征向量绘制在图上:
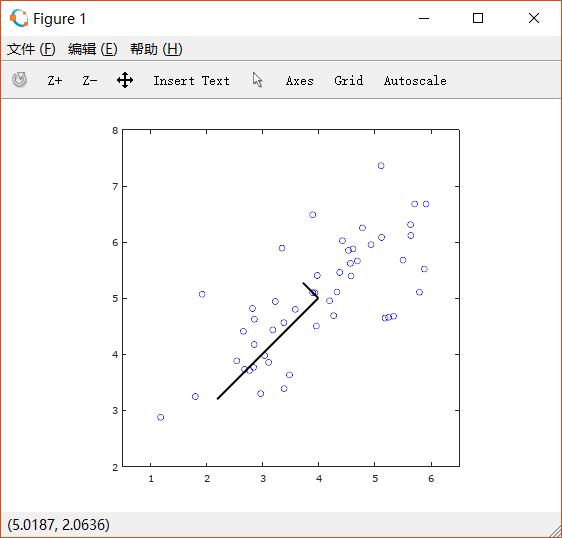
2.3 Dimensionality reduction with PCA
将高维的examples投影到低维上。
2.3.1 Projecting the data onto the principal components
完成projectData.m如下:
function Z = projectData(X, U, K)%PROJECTDATA Computes the reduced data representation when projecting only%on to the top k eigenvectors% Z = projectData(X, U, K) computes the projection of% the normalized inputs X into the reduced dimensional space spanned by% the first K columns of U. It returns the projected examples in Z.%% You need to return the following variables correctly.Z = zeros(size(X, 1), K);% ====================== YOUR CODE HERE ======================% Instructions: Compute the projection of the data using only the top K% eigenvectors in U (first K columns).% For the i-th example X(i,:), the projection on to the k-th% eigenvector is given as follows:% x = X(i, :)';% projection_k = x' * U(:, k);%Ureduce = U(:,1:K);Z = X * Ureduce;% =============================================================end
将X投影到K维空间上。
2.3.2 Reconstructing an approximation of the data
从投影过的低维恢复高维:
function X_rec = recoverData(Z, U, K)%RECOVERDATA Recovers an approximation of the original data when using the%projected data% X_rec = RECOVERDATA(Z, U, K) recovers an approximation the% original data that has been reduced to K dimensions. It returns the% approximate reconstruction in X_rec.%% You need to return the following variables correctly.X_rec = zeros(size(Z, 1), size(U, 1));% ====================== YOUR CODE HERE ======================% Instructions: Compute the approximation of the data by projecting back% onto the original space using the top K eigenvectors in U.%% For the i-th example Z(i,:), the (approximate)% recovered data for dimension j is given as follows:% v = Z(i, :)';% recovered_j = v' * U(j, 1:K)';%% Notice that U(j, 1:K) is a row vector.%Ureduce = U(:, 1:K);X_rec = Z * Ureduce';% =============================================================end
2.3.3 Visualizing the projections
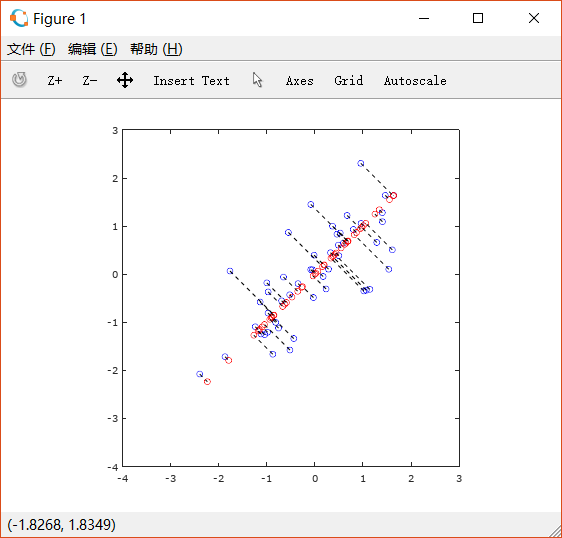
根据上图可以看出,恢复后的图只保留了其中一个特征向量上的信息,而垂直方向的信息丢失了。
2.4 Face image dataset
对人脸图片进行dimension reduction。ex7faces.mat中存有大量人脸的灰度图() , 因此每一个向量的维数是 。
如下是前一百张人脸图:
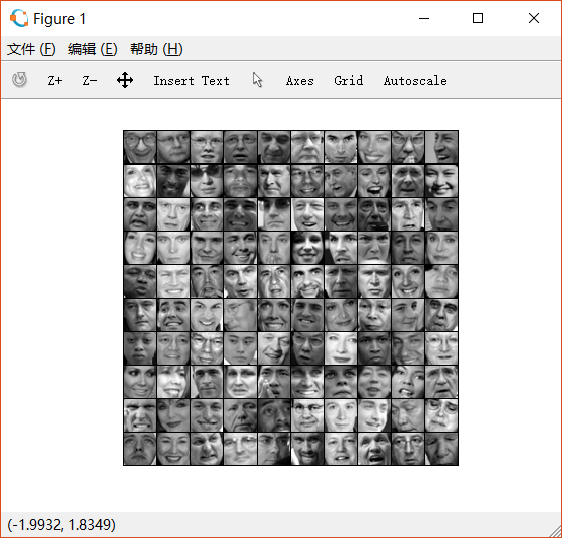
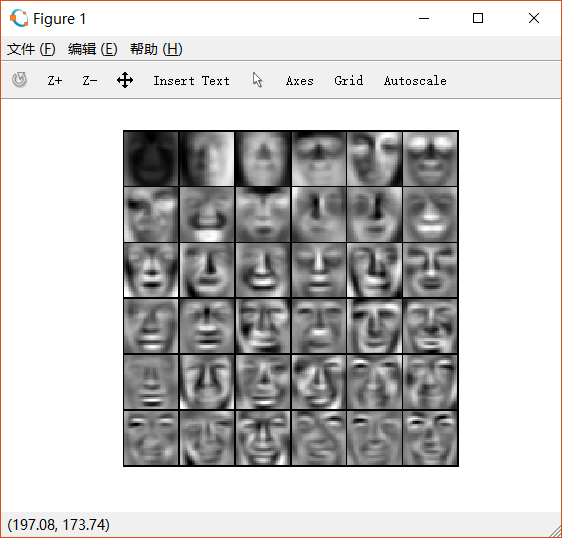
2.4.1 PCA on faces
用PCA得到其主成分,将其重新转化为 的矩阵后,对其可视化,如下:(只展示前36个)
2.4.2 Dimensionality reduction
取前100个特征向量进行投影,

可以看出,降低维度后,人脸部的大致框架还保留着,但是失去了一些细节。这给我们的启发是,当我们在用神经网络训练人脸识别时,有时候可以用这种方式来提高速度。
2.5 Optional (Ungraded) exercise: PCA for visualization
PCA常用于高维向量的可视化。
如下图,用K-means对三维空间上的点进行分类。
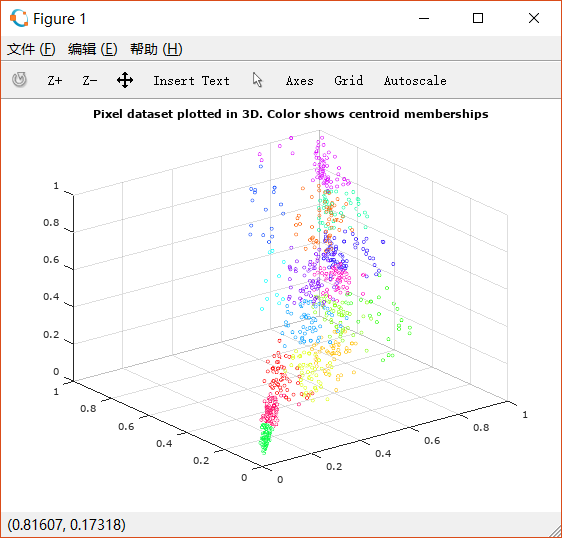
对图片进行旋转,可以看出这些点大致在一个平面上

因此我们使用PCA将其降低到二维,并观察散点图:
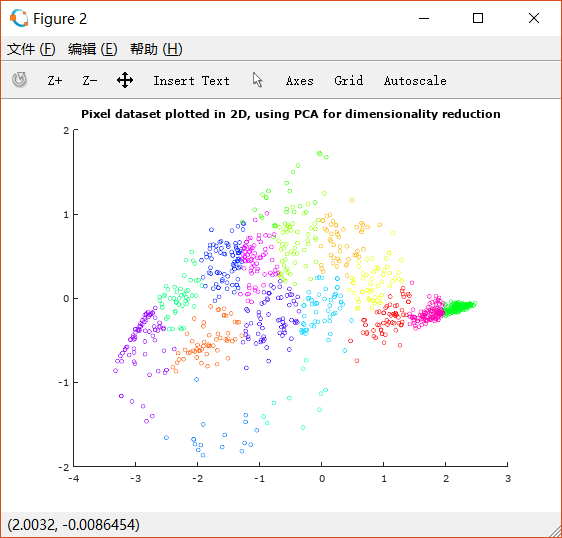
这样就更利于观察分类的情况了。
