@Billy-The-Crescent
2019-10-15T03:30:27.000000Z
字数 12107
阅读 607
Comptational Structural Biology
iGEM Modeling SCU-China
Introduction
In the metabolic circuit of cordycepin and pentostatin, the three key enzymes called cns1, cns2 and cns3 are first discovered in 2017 [Xia Y et al. (2017)], and they are poorly studied, with no structures published, which makes our project a black box without knowing the catalytic mechanism and blocks the further developement of enzyme engineering conducting on them. Based on this situation, we constructed structural prediction, molecular dynamic simulation to enrich the characterisation of the structural properties of the enzymes step by step. Additionally, we use molecular dynamic simulation and molecular docking to help determine which linker of the fusion expression proteins is prior to choose for experiments.
Structure Prediction
Having only the sequences of the enzyme cns1, cns2 and cns3, the first thing to do is to build the three-dimensional structures of the enzymes. With no access to either of the methods for protein structural analysis, including Nuclear magnetic resonance (NMR), x-ray crystallography and cryo-electron microscopy (Cryo-EM), we turned to the use of structure prediction.
Homology Modeling
Protein structure prediction is a mapping from the amino acid sequence of a protein to the three-dimensional structure. It is highly important in medicine and biotechnology although it is not as accurate as the structure from the experiments. It is observed that similar sequences often adopt similar protein structures, which forms the foundation of homology modeling.
As the most popular type of protein structure simulation method, we constructed homology modeling based on servers and softwares including SWISS-MODEL [Waterhouse et al. (2018)] and Modeller Webb B et al. (2014).
Using SWISS-MODEL, we found that the tempelate coverage of cns1, cns2 and cns3 is 41%, 76%, and 26%. The result of homology modeling is accurate when the template coverage if above 60%, and is not reliable when the template coverage below 30%.
| protein | coverage | assessment of coverage |
|---|---|---|
| cns1 | 41% | fairly reliable |
| cns2 | 76% | reliable |
| cns3 | 26% | not reliable |
Considering cns3 protein, we use Modeller to try again. The template searching process failed with the threshold E-value of 10, which is the bottom line of the protein sequences. The result shows that Modeller can not find the aligned template in protein structure database which is in the range of the threshold.
Evaluating the model of cns2 reveals the low GMQE (0.50) and QMEAN Z-score(-5.9), which indicate the low quality of the result model. Likewise, the low GMQE (0.23) and QMEAN Z-score (-6.1) indicate that the result is bad.
GMQE (Global Model Quality Estimation) is a quality estimation which combines properties from the target–template alignment and the template search method. The resulting GMQE score is expressed as a number between 0 and 1. Higher numbers indicate higher reliability.
The QMEAN Z-score indicates whether the QMEAN score of the model is comparable to what one would expect from experimental structures of similar size. QMEAN Z-scores around zero indicate good agreement between the model structure and experimental structures of similar size. Scores of -4.0 or below are an indication of models with low quality.
In conclusion, our attemptation of using homology modeling to get the structure information of the enzymes failed as the low quality of the result. We will try other methods to predict the structure of the other two proteins.
Fold Recognition
Being blocked by the bad struture prediction result of cns1 and cns3, we consult Prof. Cao Yang for other methods of structure prediction. He recommended a server called ITASSER [Yang J et al. (2014)] which is a powerful structure prediction tool based on fold recognition. More details about our consultation with Prof. Cao.
Fold recognition is another method of protein modeling. It differs from the homology modeling method of structure prediction as it is used for proteins which do not have their homologous protein structures deposited in PDB, whereas homology modeling is used for those proteins which do. From its definition, we know that it solves our problem quite well, because we can not find any fair template for cns1 and cns3.
The result of the prediction by ITASSER is as follows:
| protein | C-score |
|---|---|
| cns1 | -3.74 |
| cns2 | 0.70 |
| cns3 | -1.10 |
The confidence of each model is quantitatively measured by C-score is typically in the range of [-5, 2], where a C-score of a higher value signifies a model with a higher confidence and vice-versa.
The resulting C-score of cns1 reveals that the prediction is not good, with fair result of cns3 and good result of cns2.
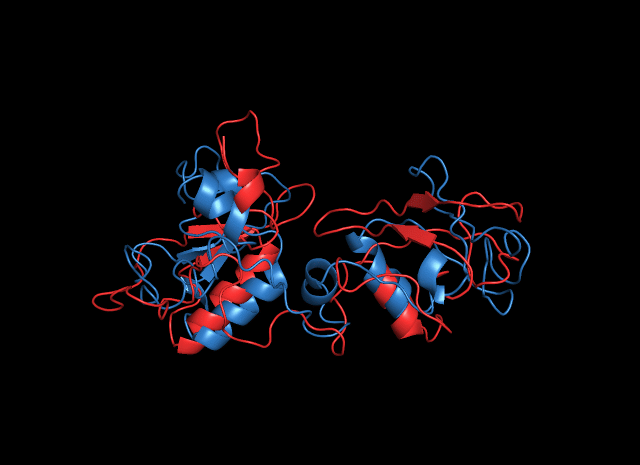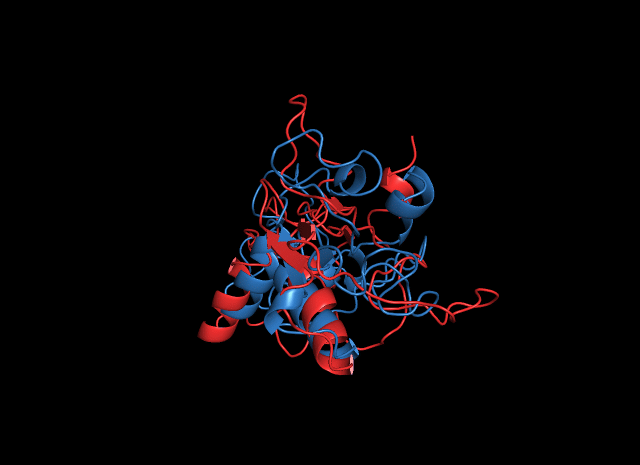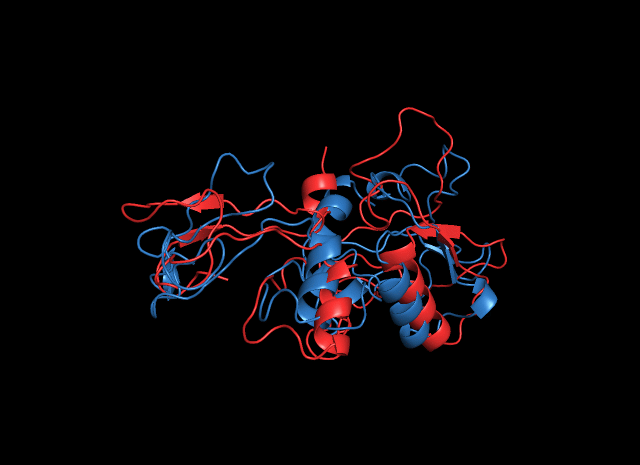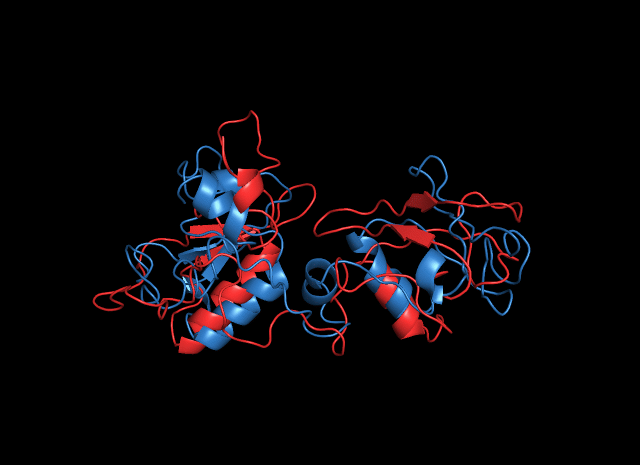
Comparing the predicted structure of cns1 by SWISS-MODEL and ITASSER, we found some interesting common points.
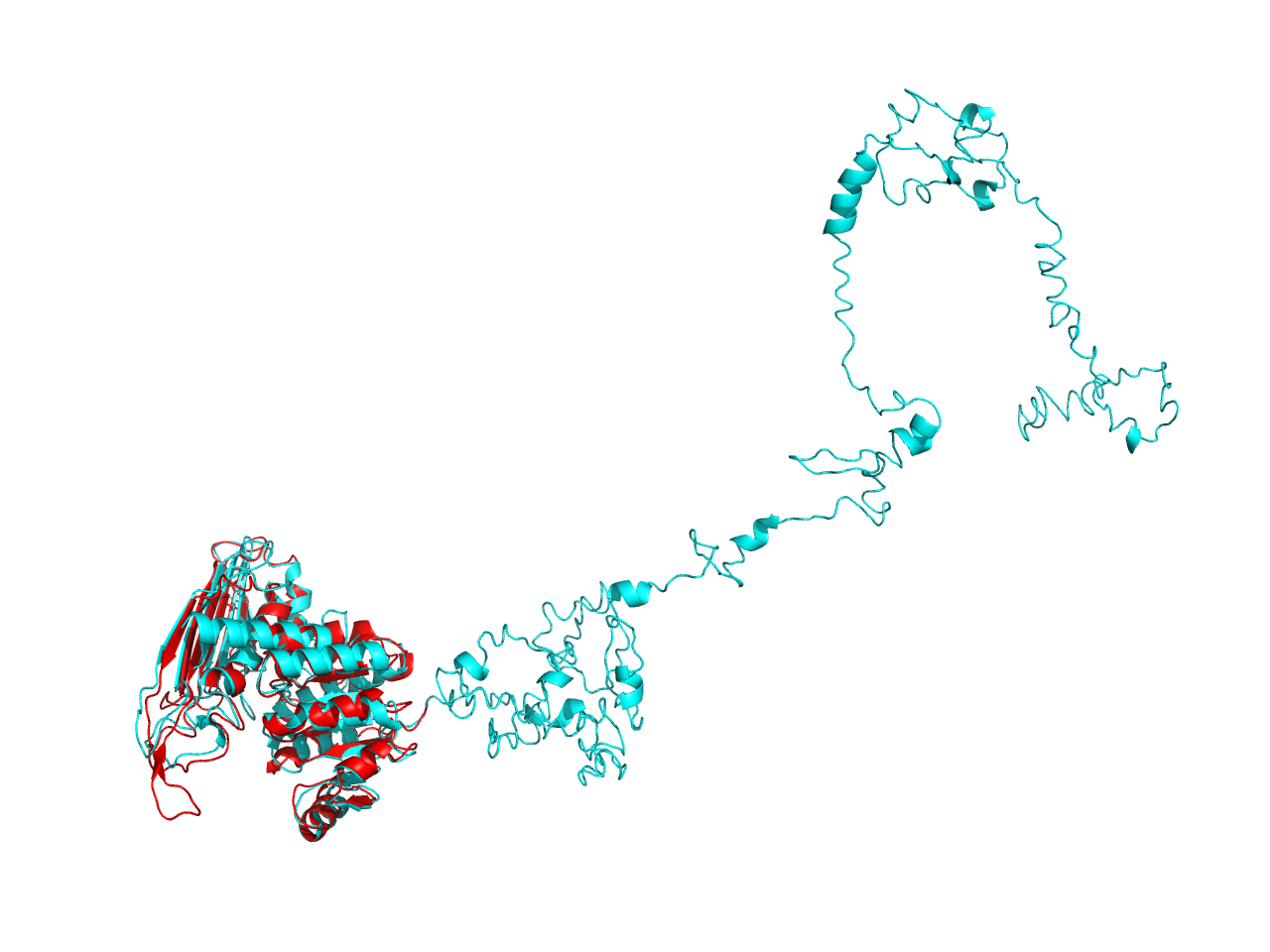
The structure from SWISS-MODEL only recognizes a proportion of the whole sequence of cns1, which forms a homo-tetramer. Alignment between the monomer and the structure from ITASSER is illustrated in the figure. The result shows that the result from ITASSER gives more information although this part of the information is not accurate (the structure in blue, aside from the alignment area, is unreliable). The two structures from both software have high similarities in the alignment area, which indicates that this area is more reliable.
Analysis of ligand binding sites by ITASSER reveals that it is located in this alignment area, which suggests that the inaccuracy of the other parts does not affect its functional characterization.
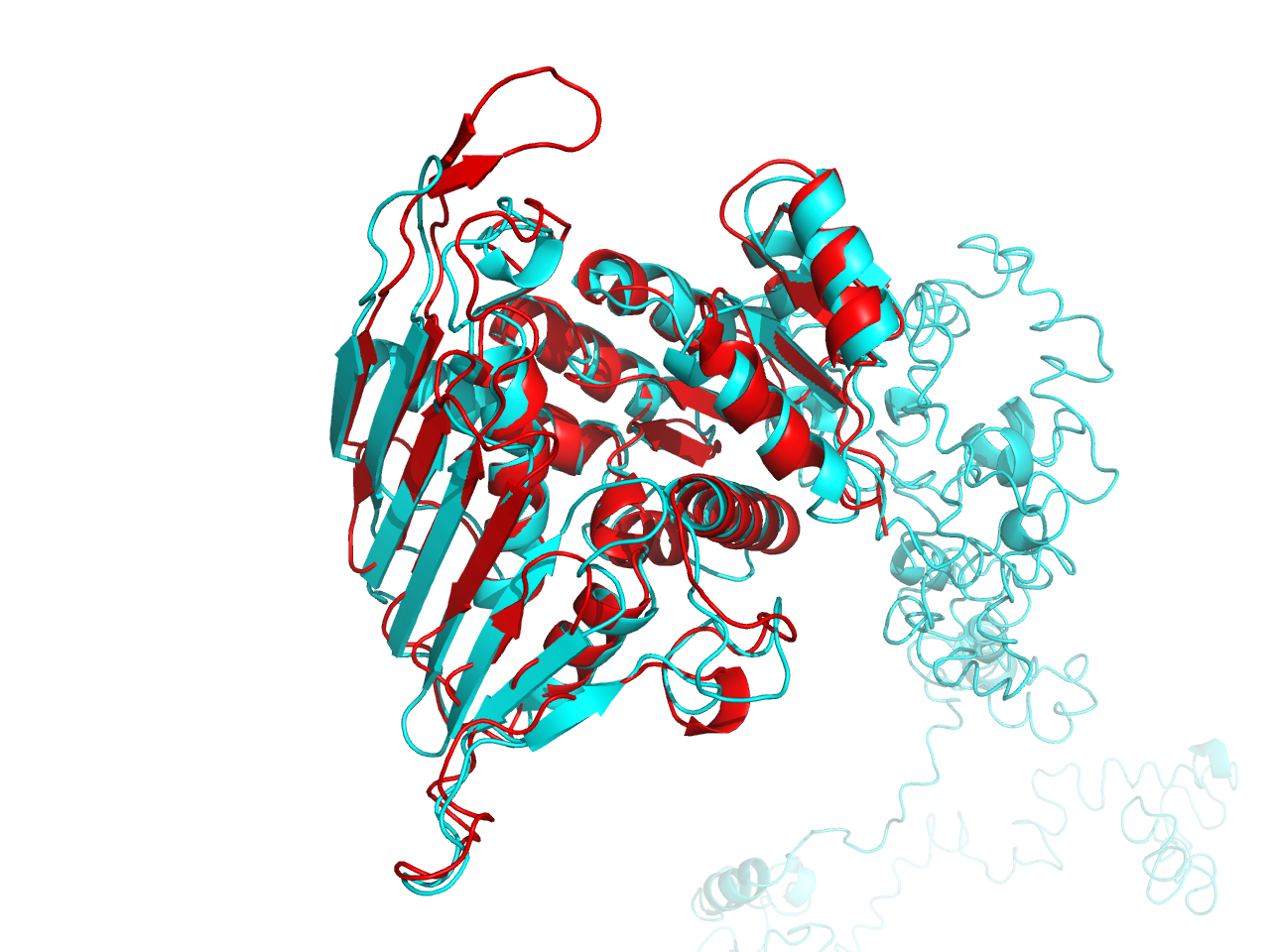
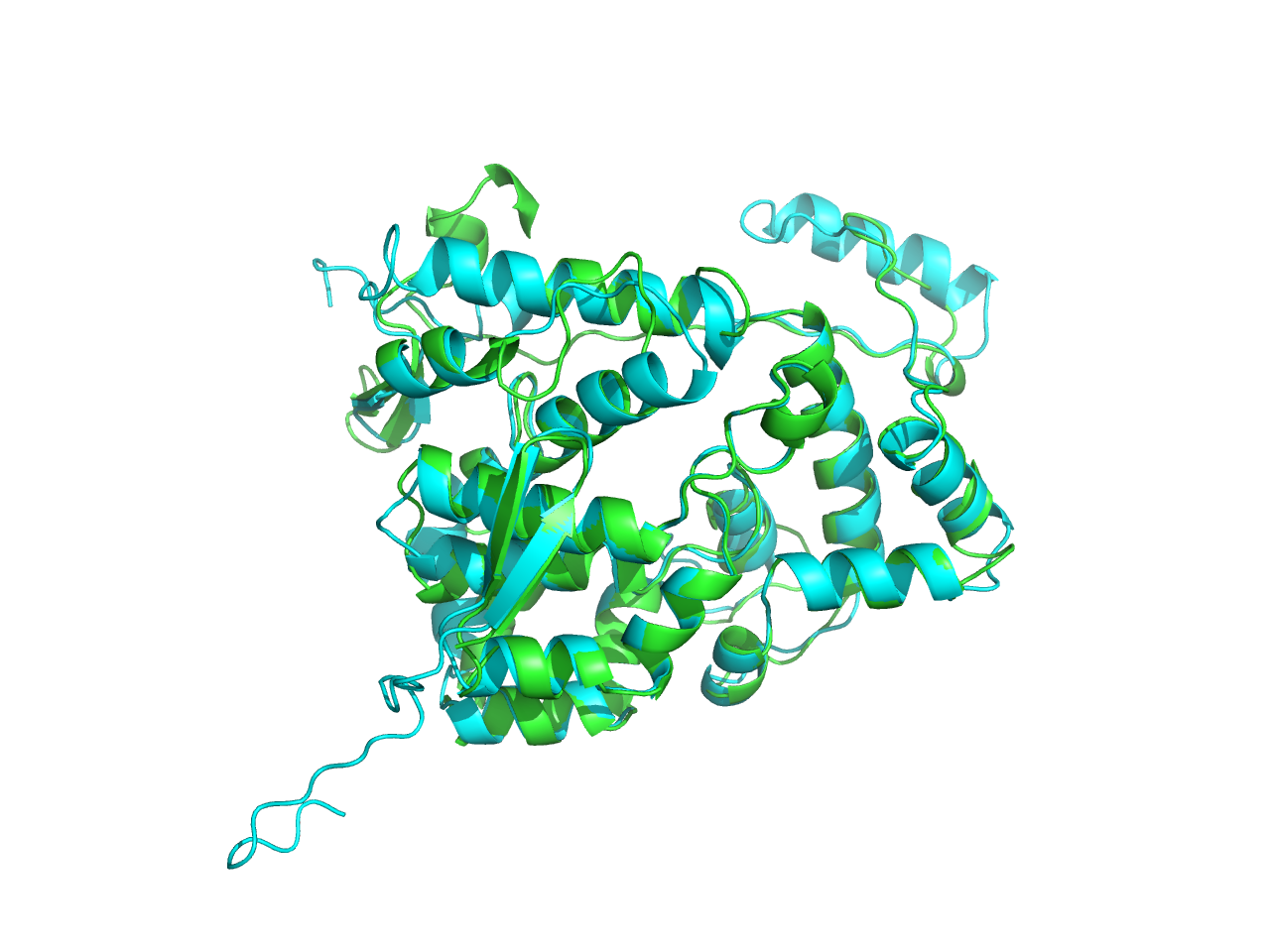
When we turned to the result of cns2 prediction, we aligned the predicted structure by ITASSER with that by SWISS-MODEL and found that the RMSD between the two is 1.492nm (aligned by Pymol), which indicates that they are quite similar. The green one is the result by SWISS-MODEL while the cyan one is the result by ITASSER.
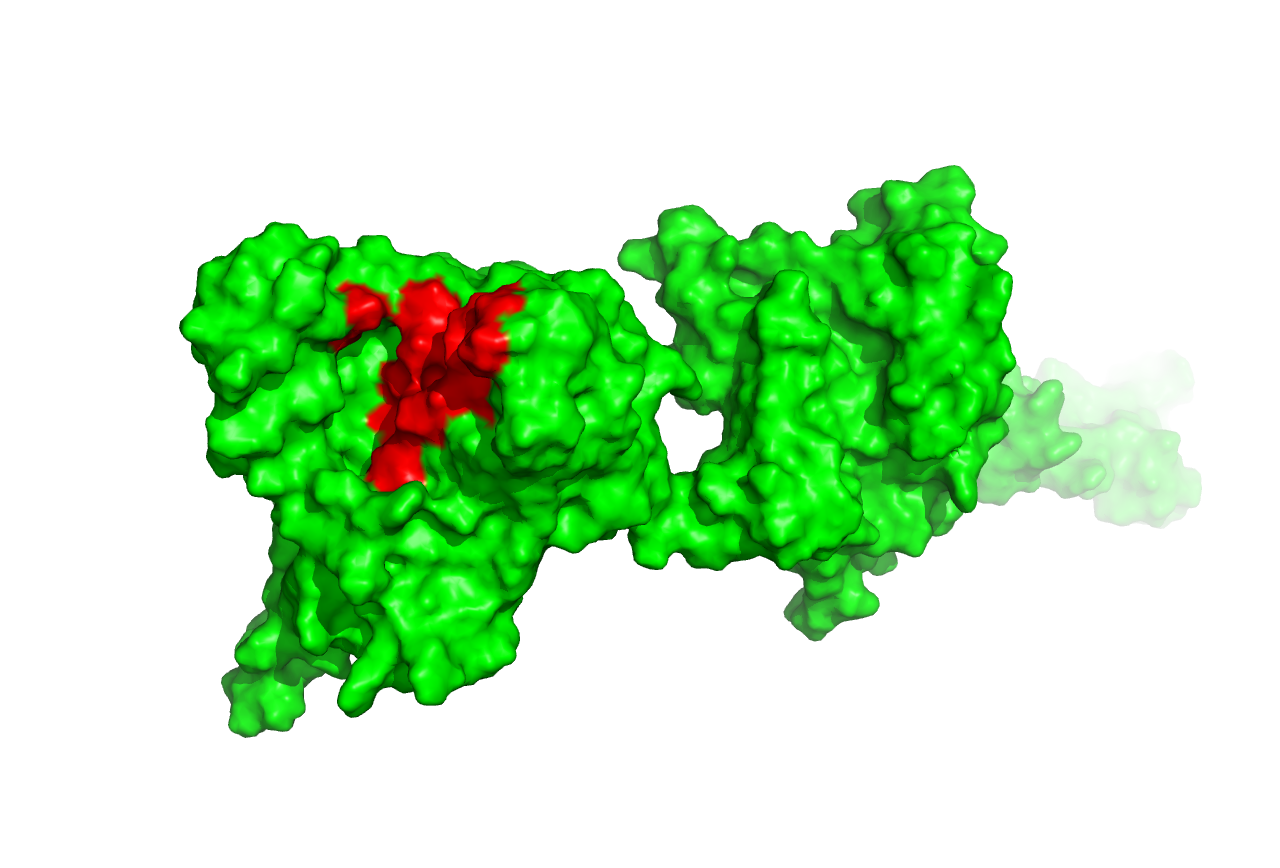
Likewise, when we analyzed the ligand-binding sites of the predicted structures by SWISS-MODEL and ITASSER when found that they are in the same position although the depth of the catalytic pocket is different. This phenomenon gave strong support to our validity of structure prediction, which did enrich the characterization of the structural properties of the enzymes.
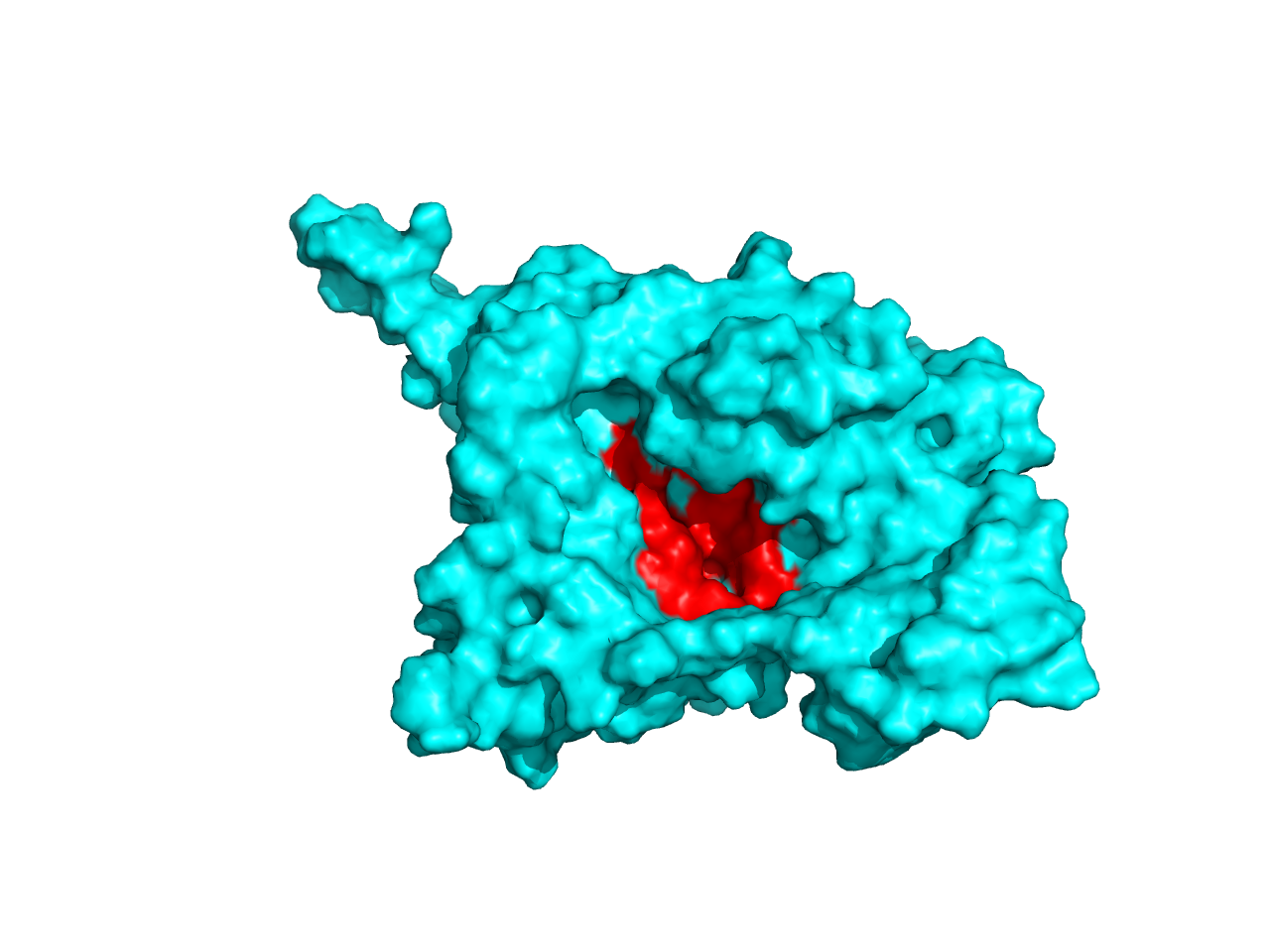
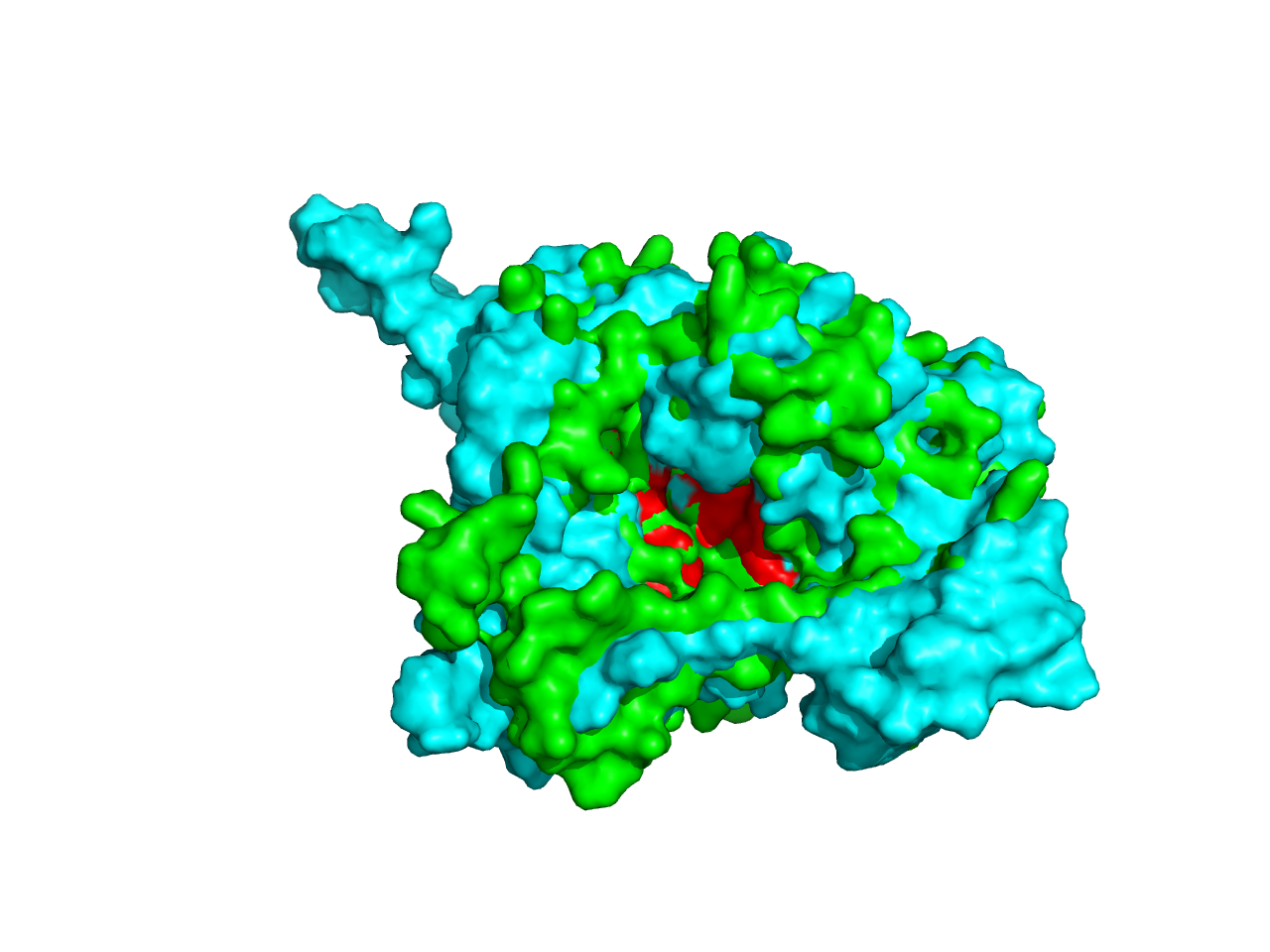
Considering cns3, the result is not acceptable, especially the structure of the HisG domain (in orange). A helix of HisG domain stretches out abruptly, which is not natural in protein. However, from the predicted result of cns3, we can attain some information. A bunch of helixes lay parallelly at the core of the protein, forming a kind of structural scaffold that supports the structural stability of NK and HisG domain. The functional domains of cns3 lay independently outside of the core. Therefore, from the angle of structural biology, in theory, separation of the two domains will not affect the individual function of each domain as they work separately. This conclusion had a theoretical guideline for the separate use of HisG domain of cns3 in our experimental design.
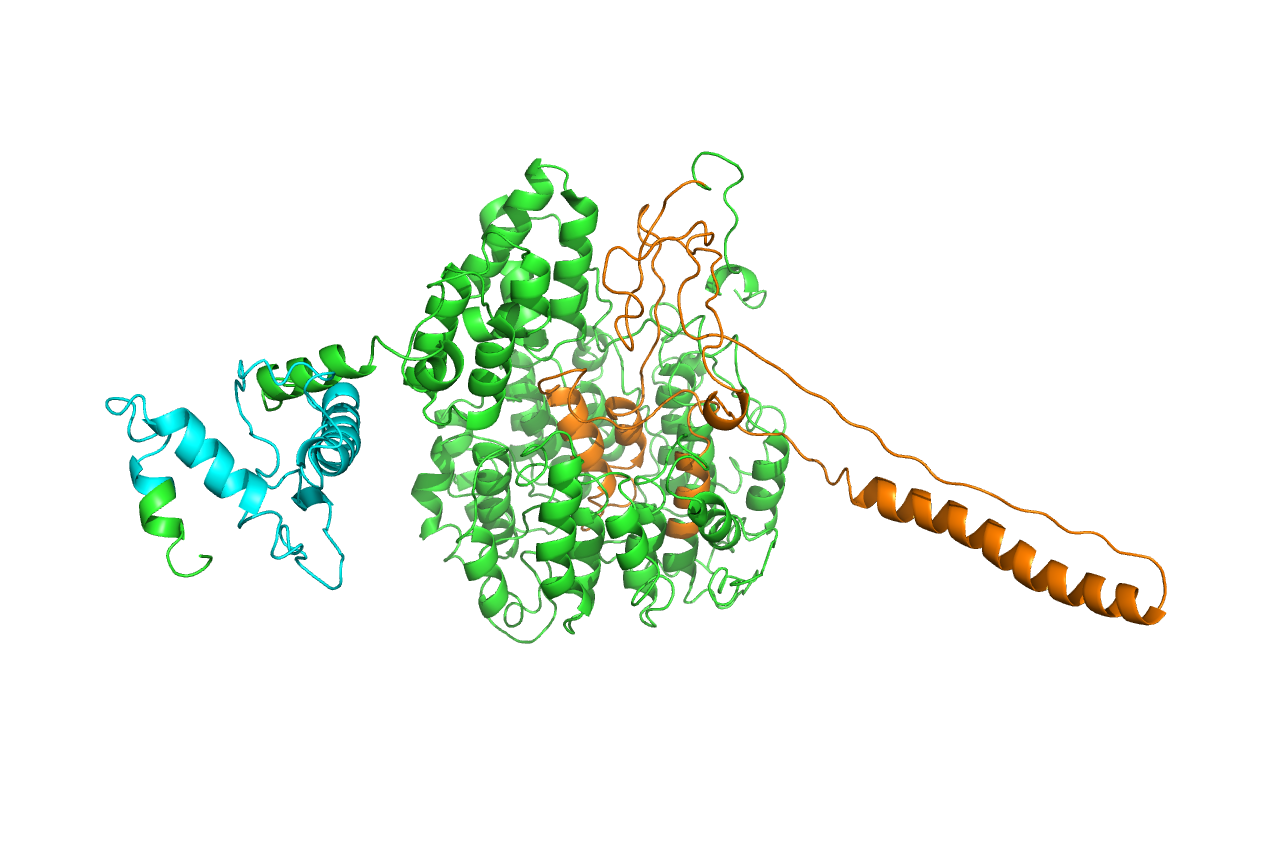
Owing to some inappropriate factors in the prediction, we further use molecular dynamic simulation to adjust the structure.
Molecular Dynamic Simulation
The prediction of the enzyme structures gave us amounts of useful information. However, it exists some improper structure in some part of the result. In order to adjust the inappropriate structure from the structure prediction process and investigate the ligand-enzyme interaction using molecular dynamics (MD) .
In our project, we use GROMACS, which is one of the most popular MD software packages for protein, lipid, and nucleic acid. The version of GROMACS we used is 5.0.7, which is run on Linux server provided by Prof. Cao. More details about the help from Prof. Cao.
Adjustment of the predicted structure
Molecular dynamics allows us to assess stability of the predicted structure of our enzymes and obtain new models that are more likely to be the native conformation in solvent which would be the possible state in our wet lab and the factory.
Steps of the utility of GROMACS are derived from the GROMACS tutorial and are summarised to SCU-China2019 GROMACS protocol which in some details is slightly different from the tutorial.
Briefly, we generated the topology of the enzyme structure, and put it in a box (the figure below), in which we filled in water and ions. We conducted energy minimization using the gradient descent method. After the energy minimization, we performed a position-restricted pre-equalization simulation and conducted the final simulation. After some time, we can get the trace file of the simulation which can be transformed into figures.

According to the GROMCAS tutorial, the final simulation after equilibrium is 1ns in totoal. Therefore, we firsly conducted the simulation for 1 nanosecond. The results for the three enzymes are as follows:
For each enzyme, the corresponding parameters including the potential energy fluctuation and temperature fluctuation are depicted below:
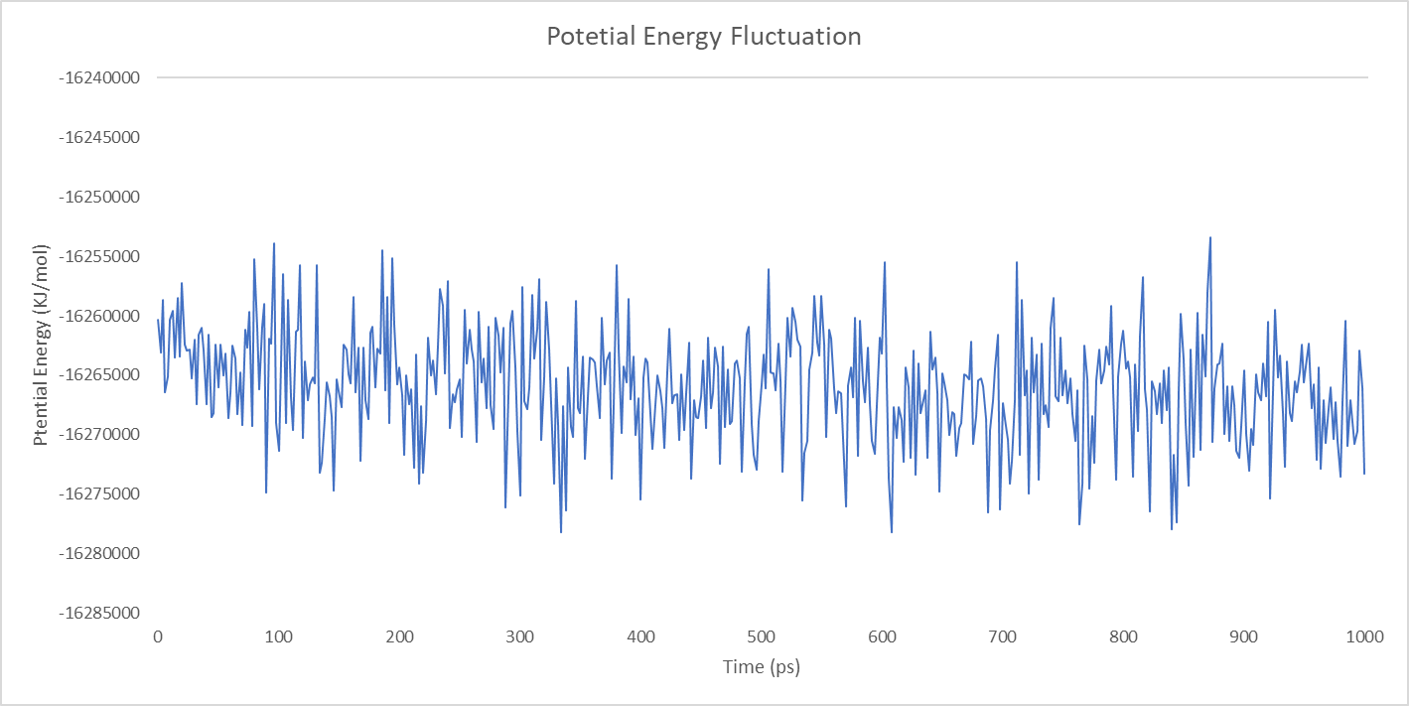
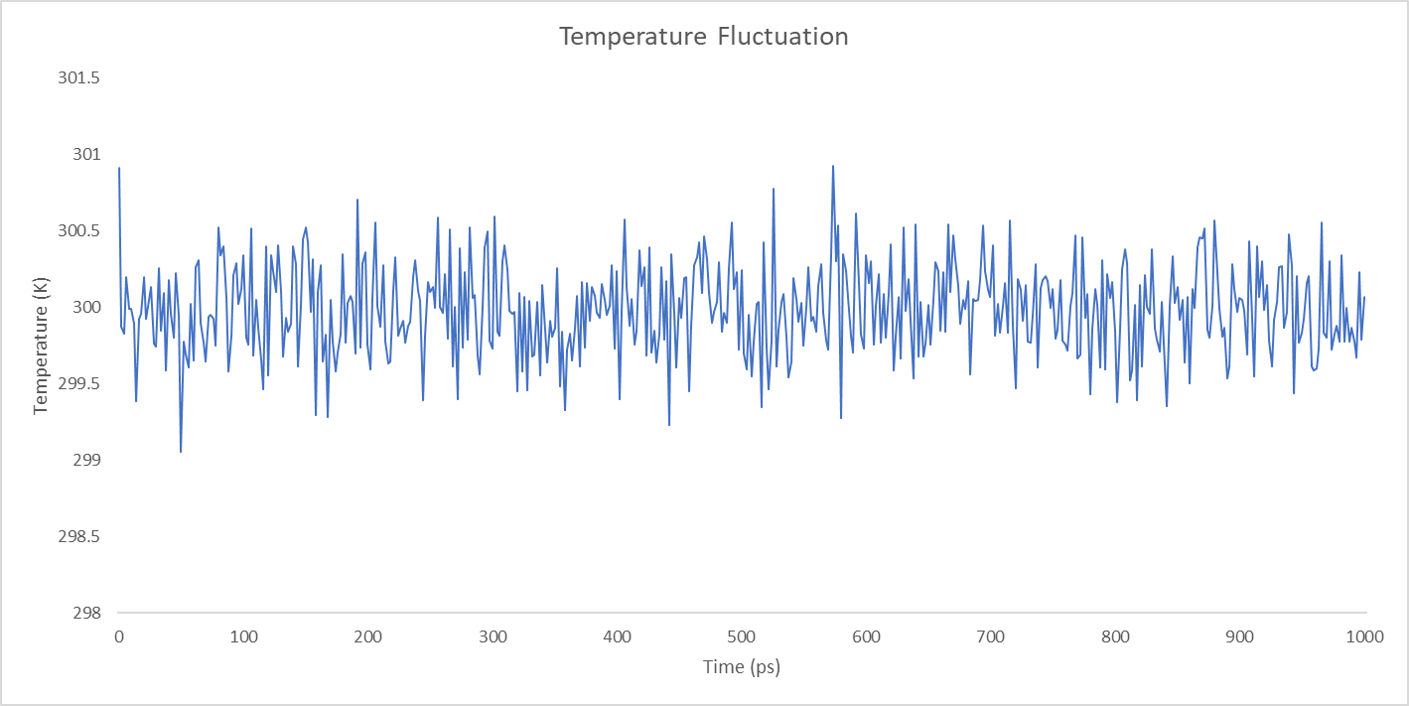
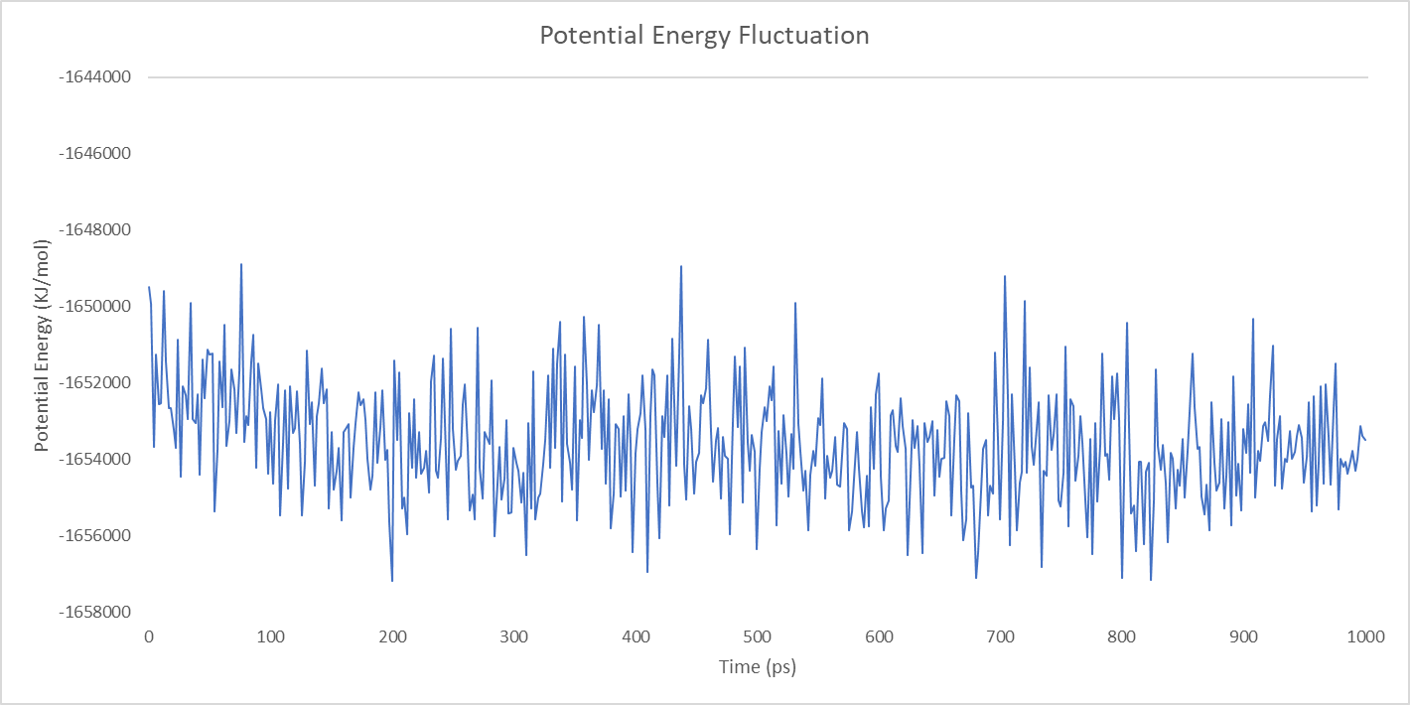
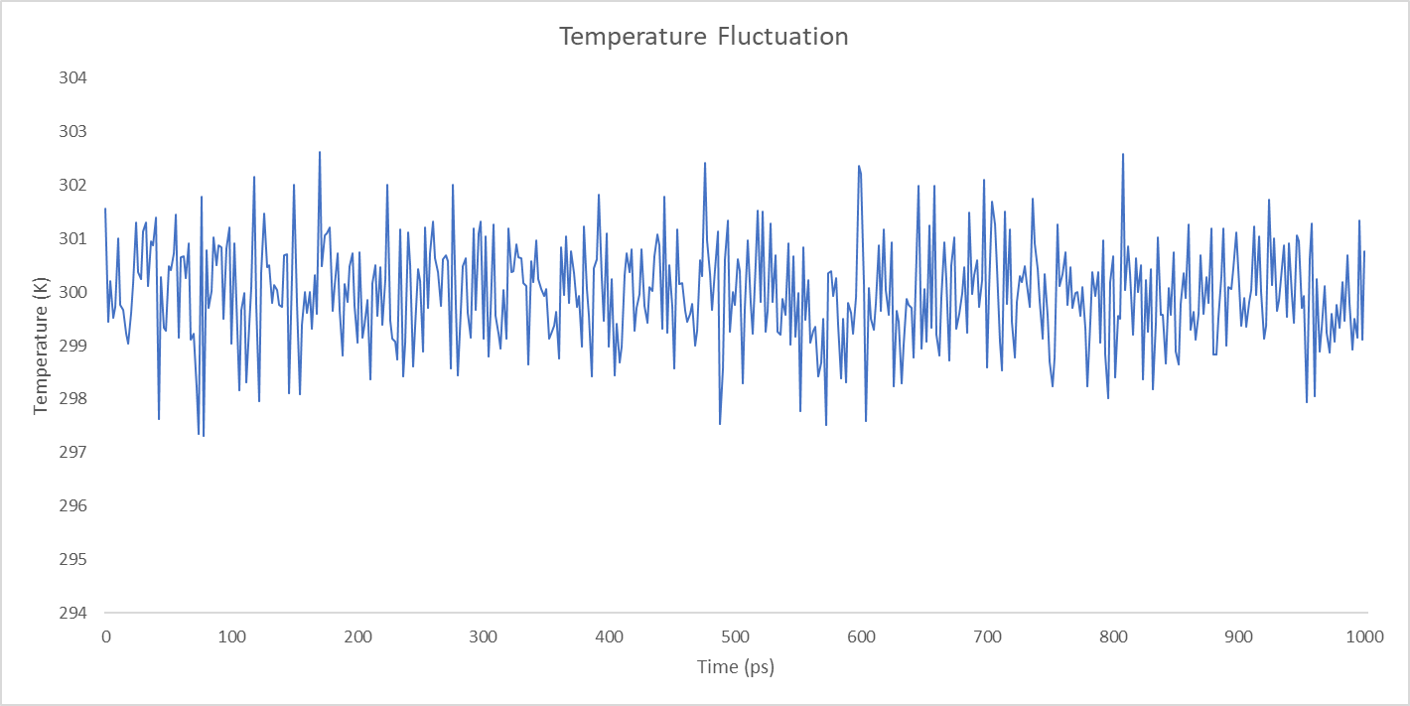
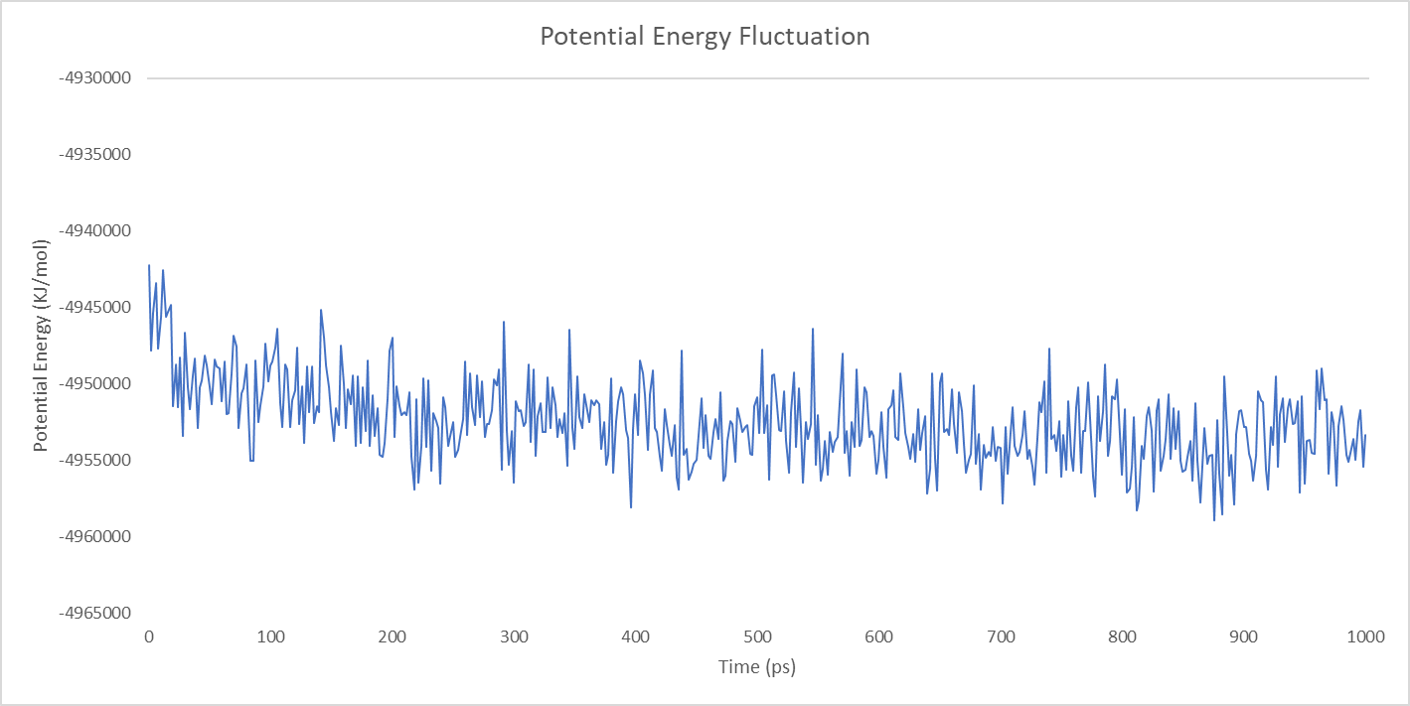
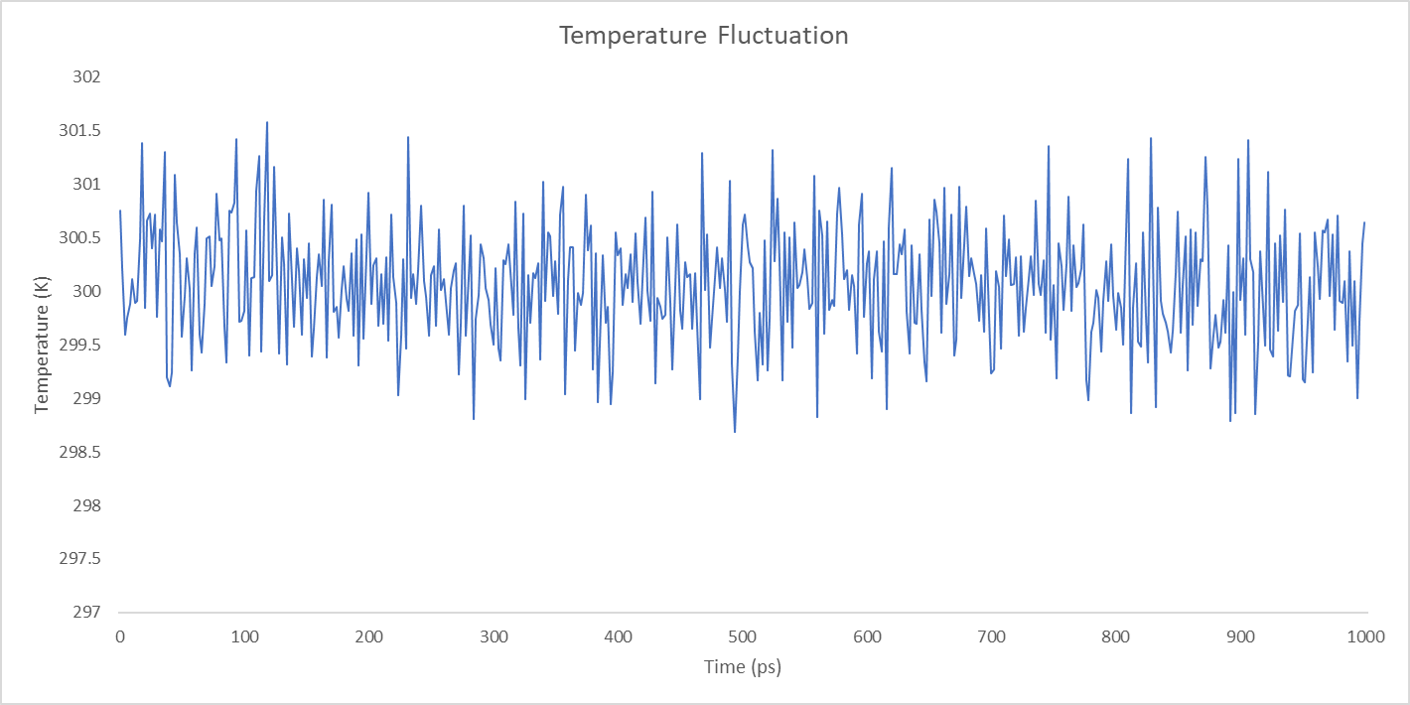
We can draw a conclusion from the results above that the structures of cns1, cns2 and cns3 are getting more stable and the average temperature in which the structures are stable is around 300K.
Alignment between the initial structures and the simulated structures will give us a straight overview of the simulation process.
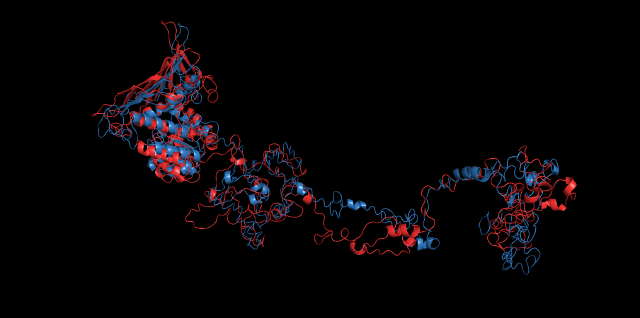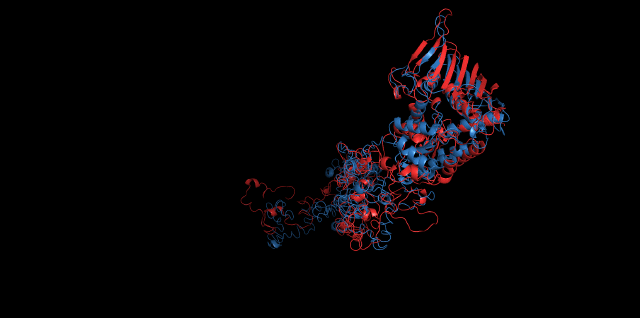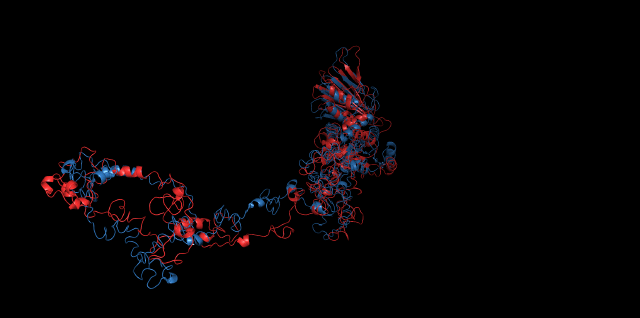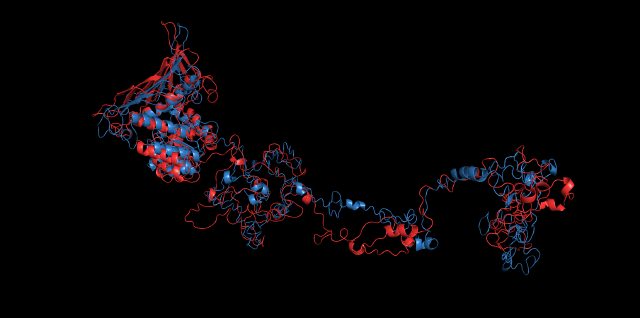
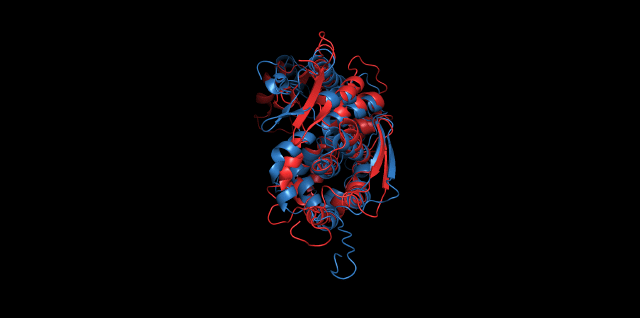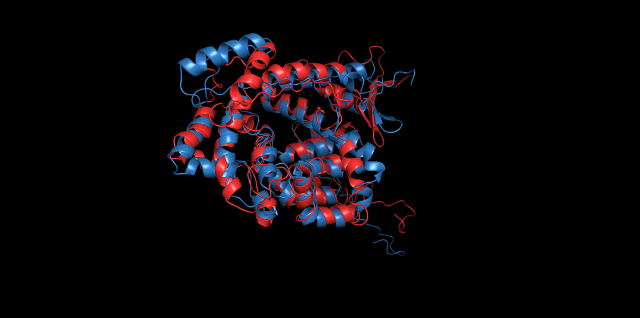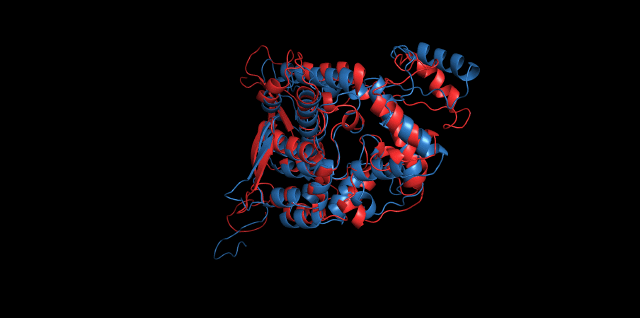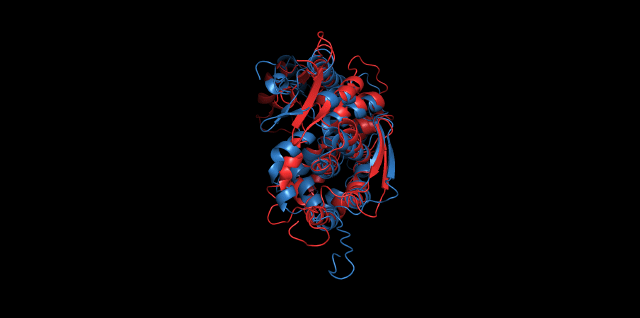
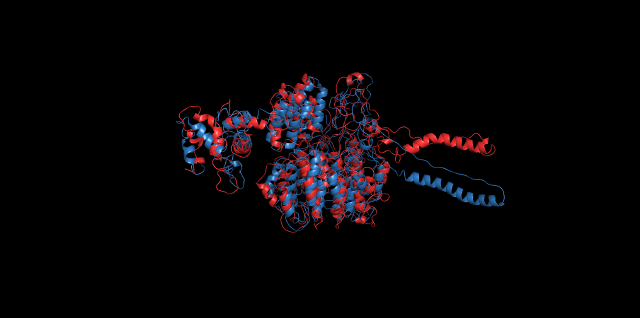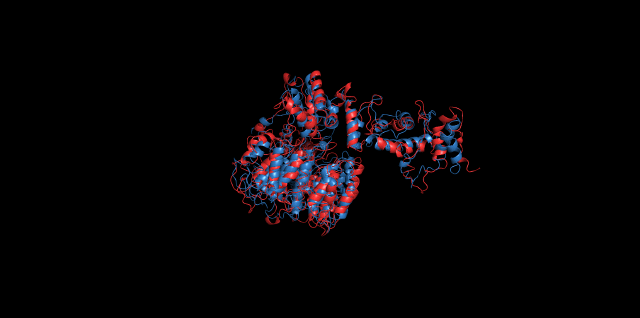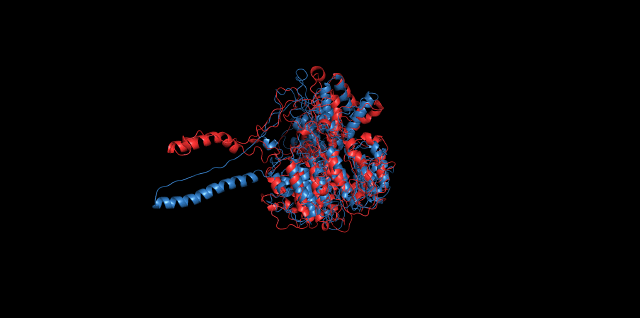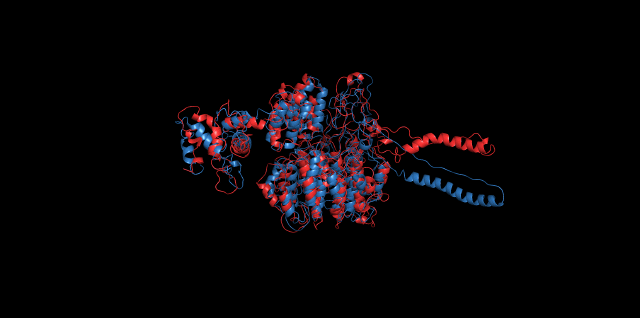
Alignment of cns1 initial structure and its simulated structure shows that it changes little in the head, which is the enzyme catalytic site while it changes a lot in its long tail. This result supports that the structure of the tail is not reliable while the head is reliable. Likewise, the analysis of cns2 reveals that this structure is reliable and the enzyme catalytic site is in its center. Interestingly, the result of cns3 shows that the structure of HisG domain changes a lot and has a tendency of curling up and folding.
Further investigation of the alignment of cns2 initial structure and its simulated structure in surface mode reveals that the core is not destroyed during molecular kinetic simulation and the catalytic pocket is shallower than the initial structure.
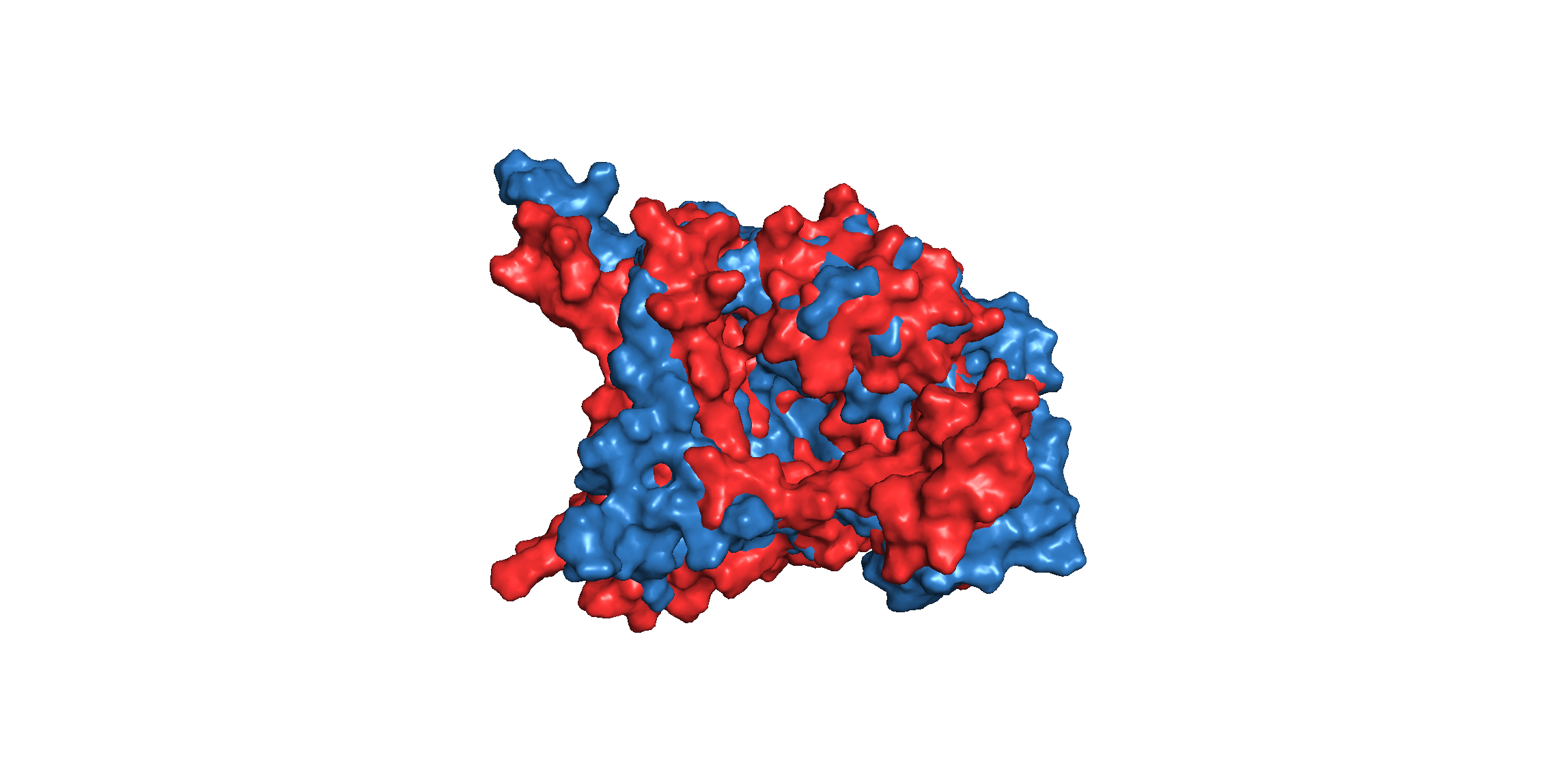
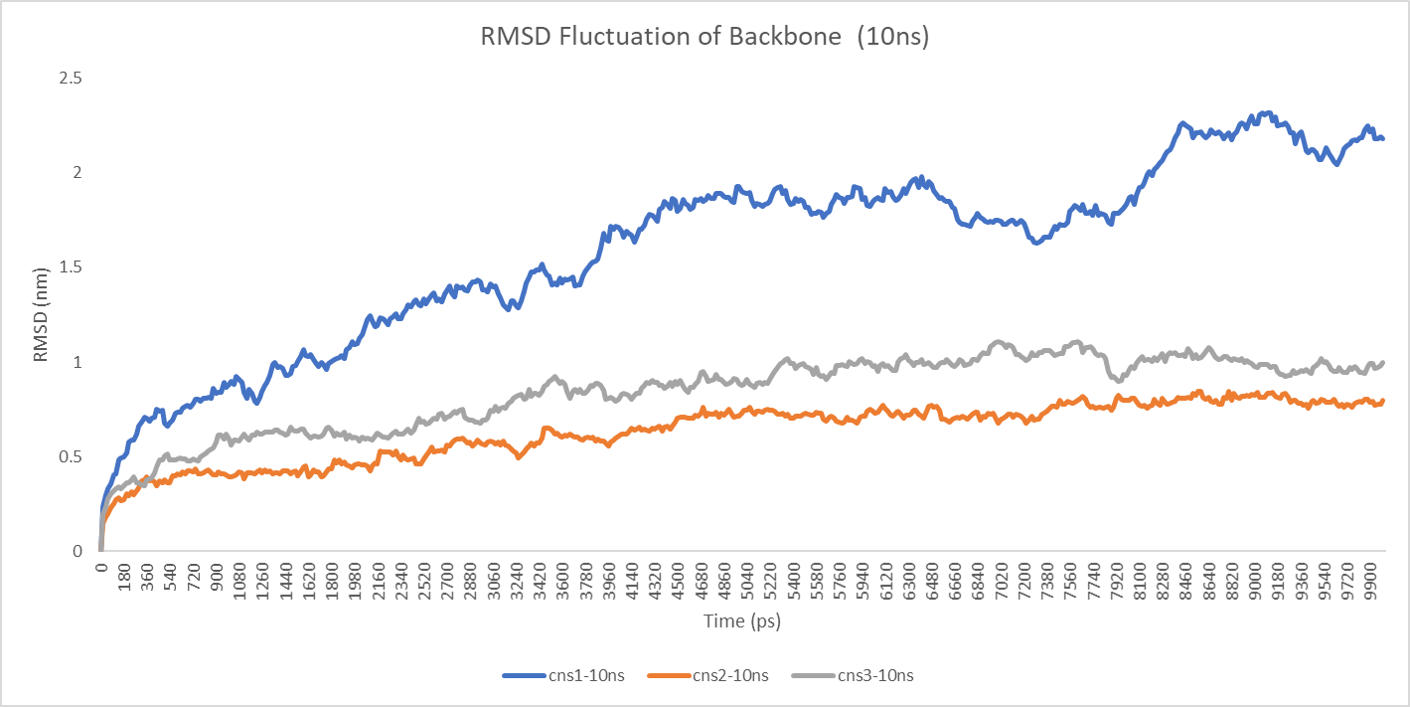
The backbone RMSD between the initial structure and the simulated structure is plotted below:
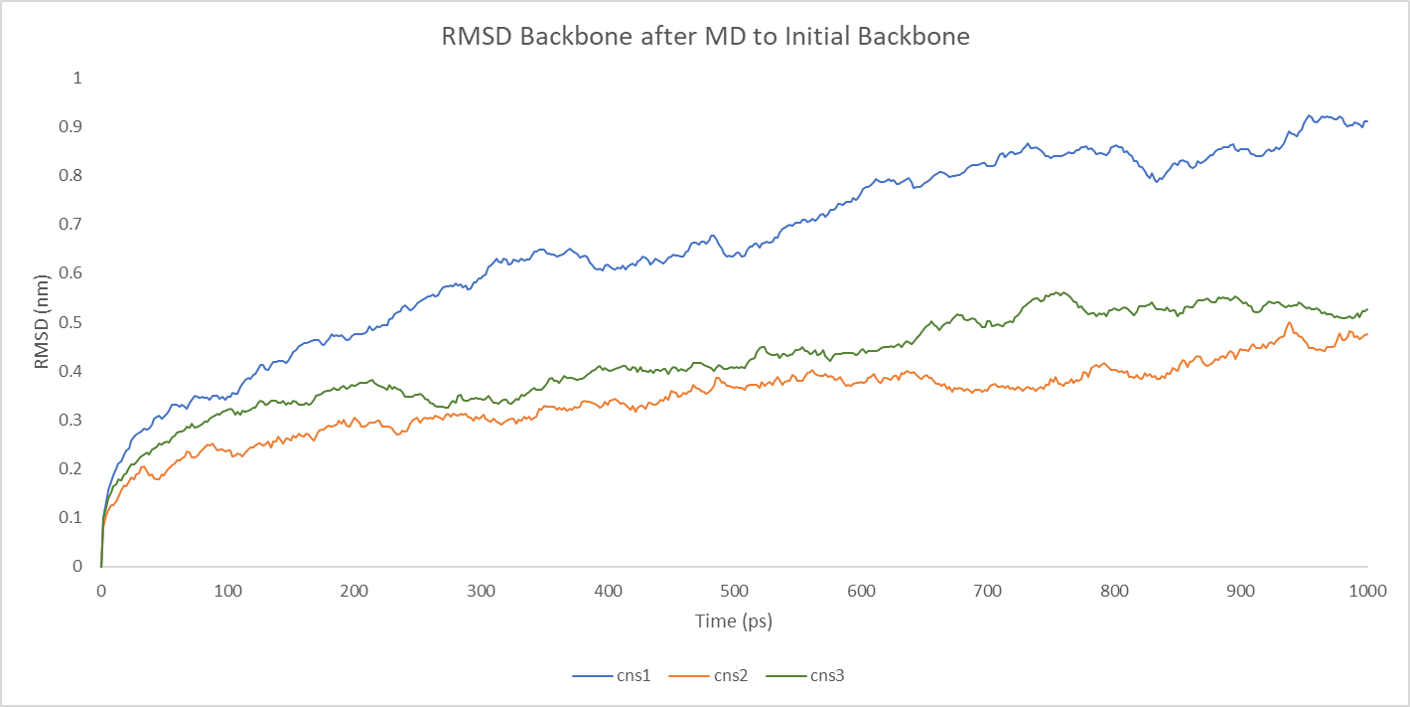
The RMSD plot above shows a fatal error in the simulation result. Overall, the RMSD lines are constantly rising with some fluctuation, which means that the result of simulation is not the final state of the MD simulation. If we extend the time of the simulation, probably, to 10ns, the final conformation would like to change to another state, a more stable and low-energy state.
Considering the problem above, we changed our protocol which is originally derived from the GROMACS tutorial and extend the time to 10ns. The result is more reliable actually, but costs much more time in return.
The 10-nanosecond simulations are shown below:
For each enzyme, the corresponding parameters including the potential energy fluctuation are depicted below:
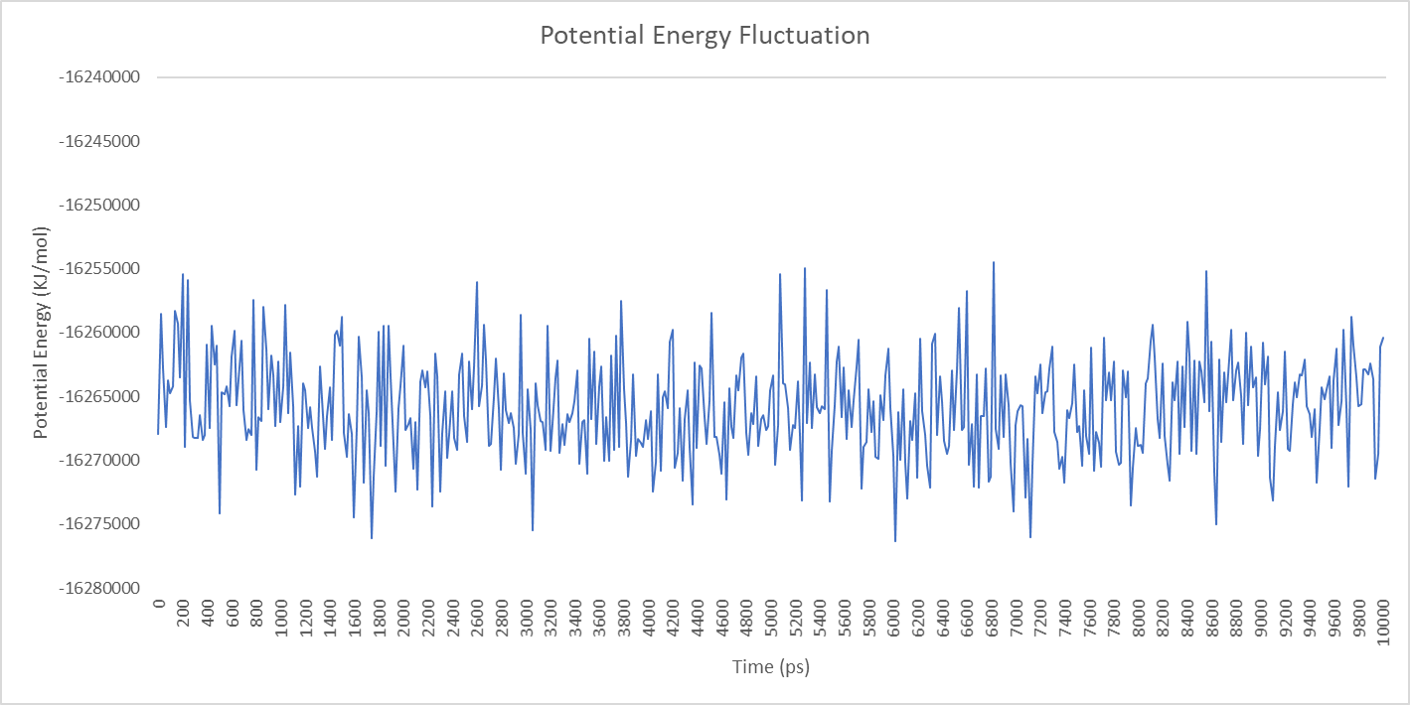
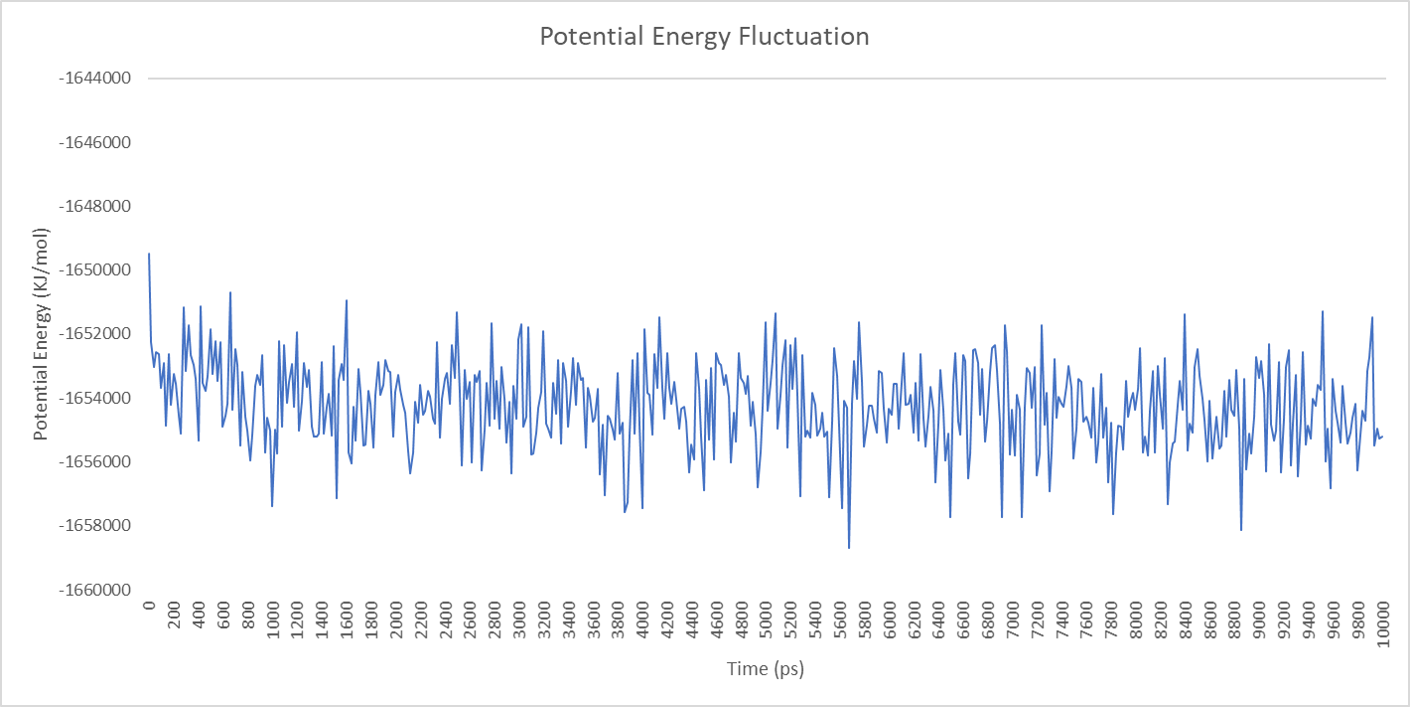
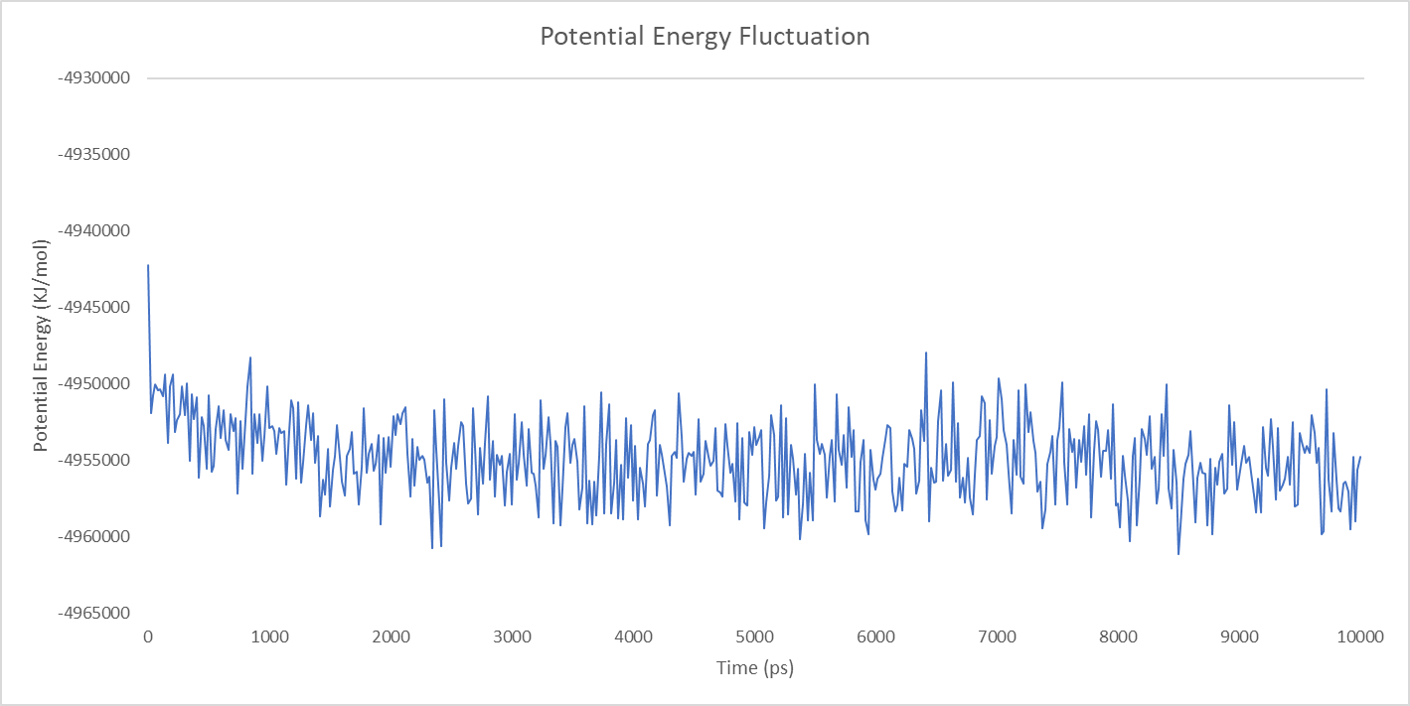
Alignment between the initial structures and the 10-nanosecond simulated structures is shown below.
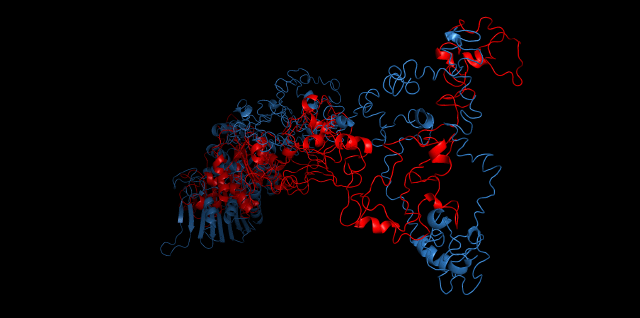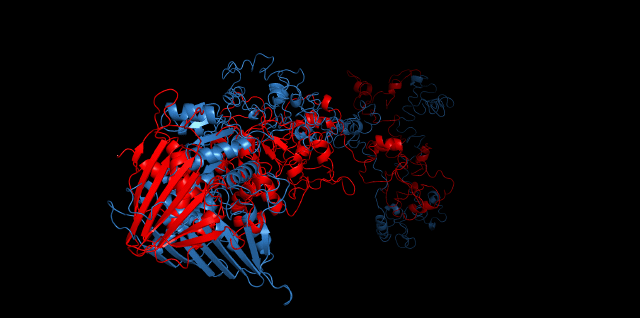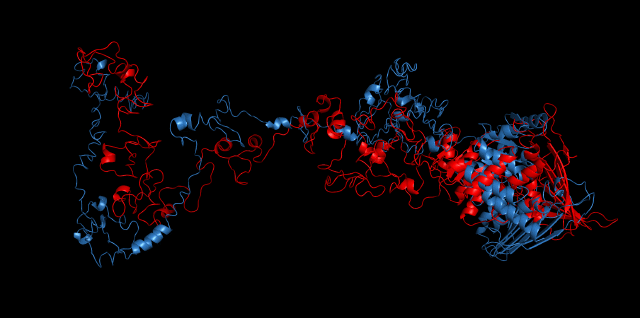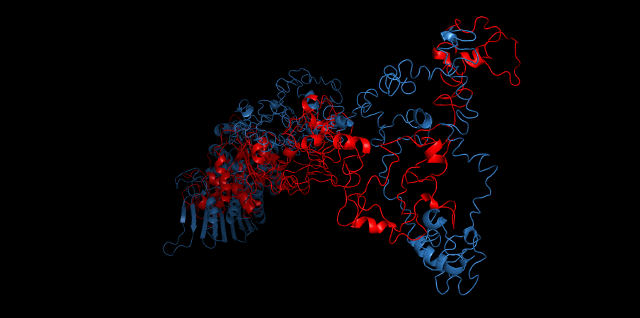
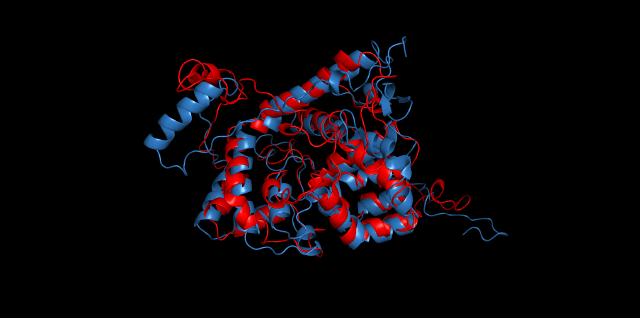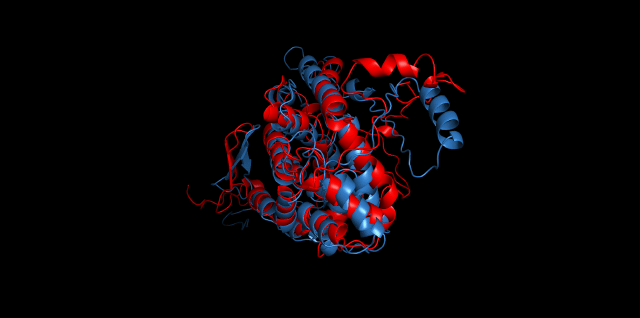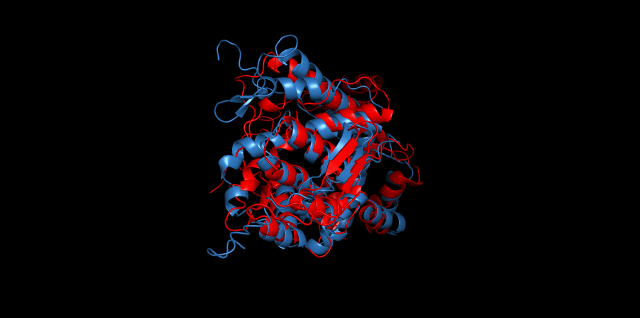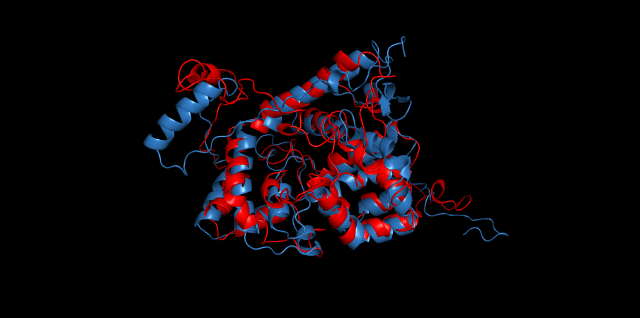
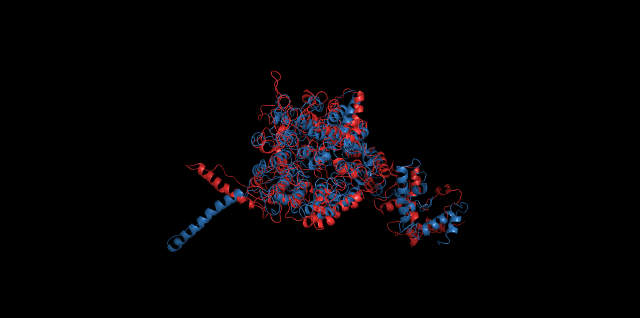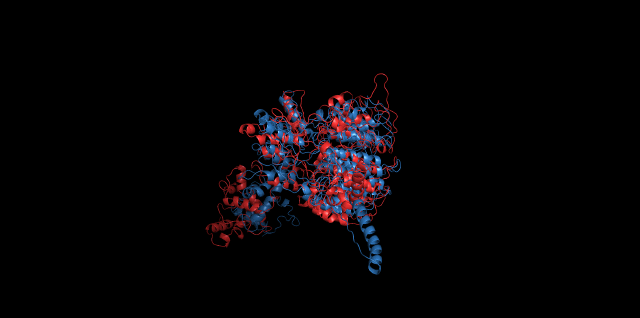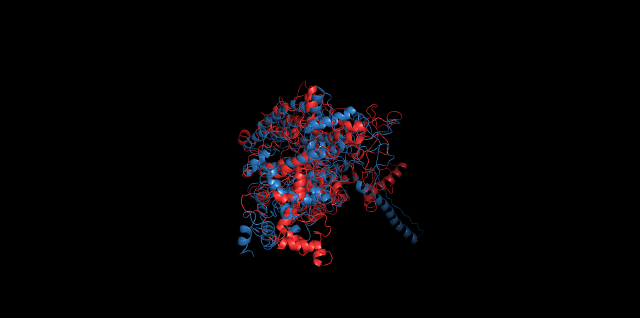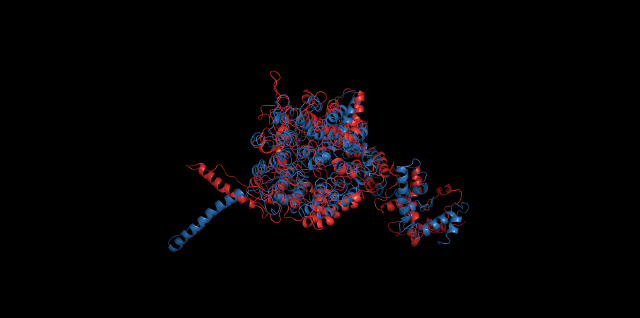
The backbone RMSD is plotted below, RMSD of cns2 and cns3 protein reaches a convergence after 10-nanosecond simulation but RMSD of cns1 does not. It seems that the simulated structure of cns1 is not the stable structure and will adjust further if time permitted. From the tendency of the structure, we can see that the red arm streching out of the core will gradually fold and adjoin to the core based on the result of 10-nanosecond simulated stricture.
Since HisG is a relatively independent domain in cns3 enzyme and it is reported in Cordyceps militaris that HisG produces pentostatin independently, it should be valid that the structure of HisG alone remains its catalytic activity. Therefore, we would like to investigate futher on the structure of HisG domain.
We performed structure prediction by ITASSER and molecular docking by SWISS-Dock for HisG. The result is below:
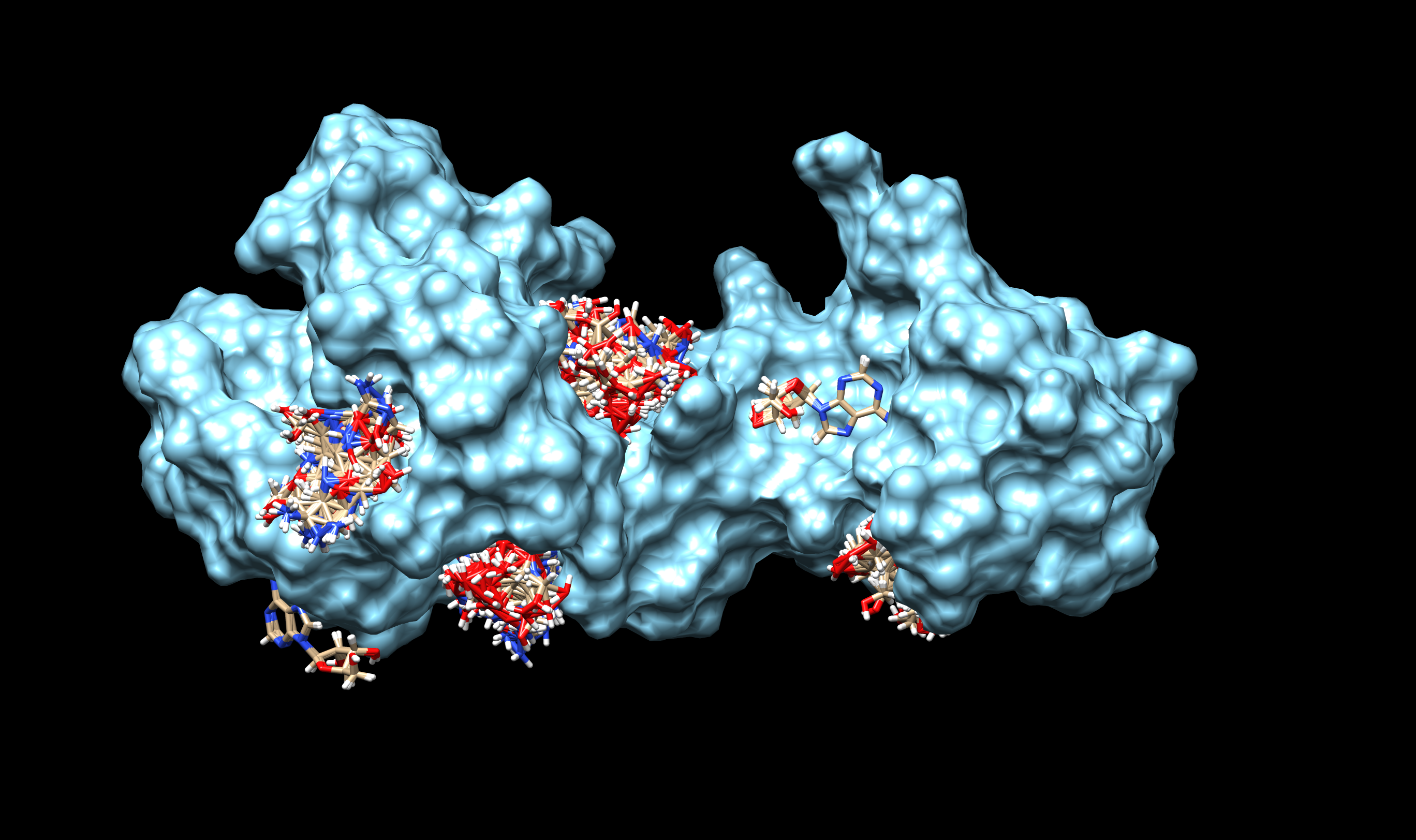
The small molecules are the potential binding conformation of adenosine, which is the substitute. It shows some probable binding sites of adenosine to HisG domain. However, this structure is not natural from the angle of structural biology because it has two huge pockets which are much bigger than they should be. Therefore, we conducted molecular dynamics simulation further for this structure of HisG. The 10-nanosecond simulation is below:
Simulated structure of HisG domain of cns3 protein shows a smaller and deeper package in catalytic core compared with the initial predicted structure. This structure convinced us that it would work well in Saccharomyces cerevisiae. More details about the function validation of HisG domain.
Linker Determination
The designing principle and evaluation of the linker between two proteins used in fusion expression are obscure, so it is challenging to design linker from scratch. Although some linkers can be found in some articles, but we have no idea if these linkers fit our project well unless doing experiments. However, the painstaking vector construction and function tests prevent us from doing so. Therefore, we would like to use comprehensive bioinformatic methods to evaluate the linkers collected from articles from the structural biological angle.
Reference
[Xia Y et al. (2017)]
Xia, Y. , Luo, F. , Shang, Y. , Chen, P. , Lu, Y. , & Wang, C. . (2017). Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin. Cell Chemical Biology, S2451945617303276.
[Waterhouse et al. (2018)]
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F.T., de Beer, T.A.P., Rempfer, C., Bordoli, L., Lepore, R., Schwede, T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46(W1), W296-W303 (2018).
[Webb B et al. (2014)]
Webb, B. , & Sali, A. . (2014). Comparative protein structure modeling using modeller. Current Protocols in Bioinformatics, 47.
[Yang J et al. (2014)]
Yang, J. , Yan, R. , Roy, A. , Xu, D. , Poisson, J. , & Zhang, Y. . (2014). The i-tasser suite: protein structure and function prediction. Nature Methods, 12(1), 7-8.
GROMACS Tutorial: http://www.mdtutorials.com/gmx/index.html
