@Billy-The-Crescent
2019-04-04T07:44:57.000000Z
字数 5302
阅读 967
iGEM 2018 TUDelft
iGEM TUDeft 2018 ADOPE gene-doping
We are fighting for honest athletes and clean sports by detecting gene doping - a rising threat to fair sport.
Gene doping is the misuse of gene therapy to enhance athletes’ performances, is one example of human enhancement
The essential purpose of their system is to detest gene engineering.
Gene Doping
A strategy doping in athlete through gene therapy.
Adding DNA into bodies to express proteins which can enhance recipients' physical ability.
After comparing different kinds of vectors for DNA doping,
Adenoviruses are seen as the most likely transfection method for gene doping at this stage.
Process

Sample Preparation
DNA from blood sample, serum and plasma
How to distinguish host genome DNA and doping DNA?
- constitutive promoter for higher levels of expression.
- The introns will be taken out, resulting in exon-exon junctions.

Why sfDNA contains doping DNA?
Injection remain.
Can serum and plasma keep the DNA for a long time?
Plasma and serum containing cfDNA can be stored frozen and stable for longer periods of time, up to years, without significantly lowering the concentration of the cfDNA.
Intergrated Modeling:

Injection of 141 billion viral vectors
The detection limit of 1000 copies per mL of blood
Target DNA:
Not one or two, but take human erythropoietin (EPO) gene as an example.
Prescreening
Intergrated Human Practice
Advice from the Dutch Doping Authorities:
He emphasised the importance of a high throughput assay that could screen thousands of athletes simultaneously.
dextrin-capped gold nanoparticles (d-AuNPs)

The whole process is placed in an increasing NaCl environment.
What the concentration of NaCl has to be chosen?

520nm Red color
620nm Green Color
The effect of single DNA to the d-AuNPs?

Concentration of single DNA probe?

So, does the system work?

Sensitive?
in a concentration ranging from 29 fM to 2.9 aM DNA using a single DNA probe in reference.
But their lower limit is 1nM.
Library Preparation
Modification from the third-generation sequencing - nanopore sequencing.

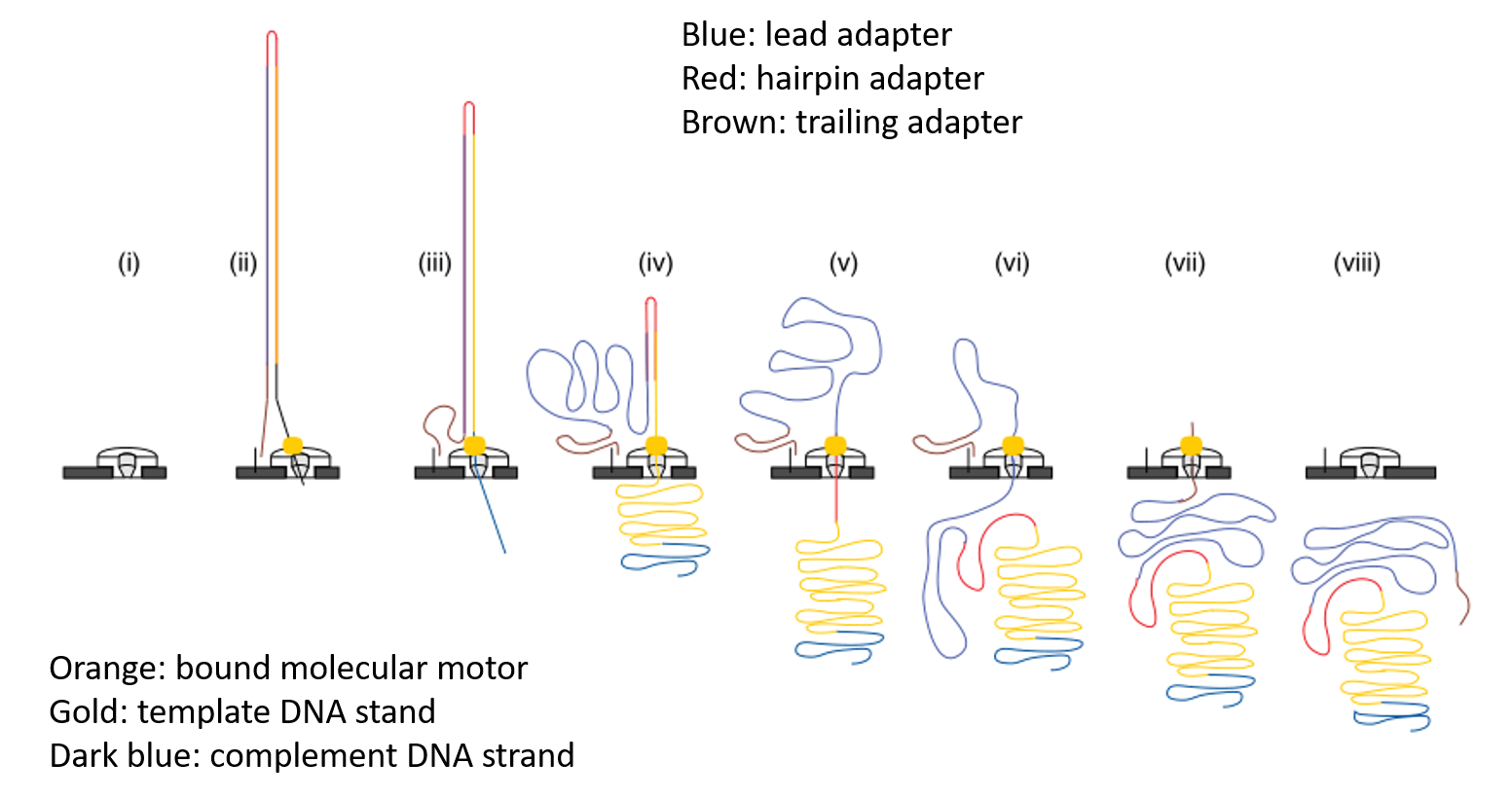
Core:
Fusion protein consisting of a Tn5 transposase and a dxCas9.

Why dxCas9?
- It has the broadest known Protospacer Adjacent Motif (PAM) sequence including NG, GAA and GAT
- It has a greater DNA specificity compared to SpCas9, resulting in significantly lower off-target activity
Tn5 transposase
This protein forms a dimer and is able to pick up specific DNA (transposon) flanked by mosaic ends (ME) sequence (CTGTCTCTTATACACATCT) and integrate it randomly into DNA.
Linker?
Glycine Helical peptide (GHP) Linker
K L G G G A P A V G G G P K A A D K
Functionality verification:
In vivo:

Two plasmids into BL21:
1. contains the fusion construct and the LacZ sgRNA cassette.
2. contains the K+ gene franked by MEs and RFP.
| Genomic | Template with RFP cotransformed? | Grown on LB+Cam+Kan | LacZ disrupted? | Colony phenotype |
|---|---|---|---|---|
| On target | No | Yes | Yes | White |
| On target | Yes | Yes | Yes | Red |
| Off target | No | Yes | No | Blue |
| Off target | Yes | Yes | No | Purple |
| None | No | No | No | N/A |
| None | Yes | Yes | No | Purple |
Do PCR and WGS for further verification.
In vitro:
Trypsin resistance assay
Loaded dxCas9 undergoes a significant conformational shift causing the protein to become significantly more
trypsin (protease) resistantcompared to an apo-dxCas9 (non sgRNA loaded) protein (Jiang et al, 2015).
We visualized this resistance to trypsin bycomparing the protein degradation ratiobetween loaded- and apo- dxCas9 on an SDS PAGE.
Electrophoretic mobility shift (binding) assay (EMSA)
A mobility shift occurs when binding of DNA to protein retards the movement of the DNA through the
polyacrylamide gel, creating two distinct bands.
Then, they use EMASA to test:
1. The fusion protein with target DNA
2. The Tn5 transposase with DNA apdapter

Intergrated Modeling
we developed a sgRNA model which identified the optimal exon-exon target site by searching for the least sgRNA's required to cover all possible variation of the target sites due to synonymous mutations of EPO coding sequence.
Result: 12 sgRNAs produced.
Innovation:
We created a barcoding webtool to generate unique barcodes, which are integrated into the adapter sequences that are ligated to the target sequence.
This allows us to sequence samples from multiple different athletes in the same run and trace the output sequence back to corresponding barcode.
Sequencing


Furthermore, add barcode between the two parts of sequence above.
Difference between ONT and their method:

Result:

The efficiency of integration was not high and some of the aligned reads showed extended regions summing up to long reads of 2 - 11 kbp.
Model Details
sgRNA model
Algorithm to generate the possible sgRNA PAM sites at exon-exon junction with least mutation possibility.
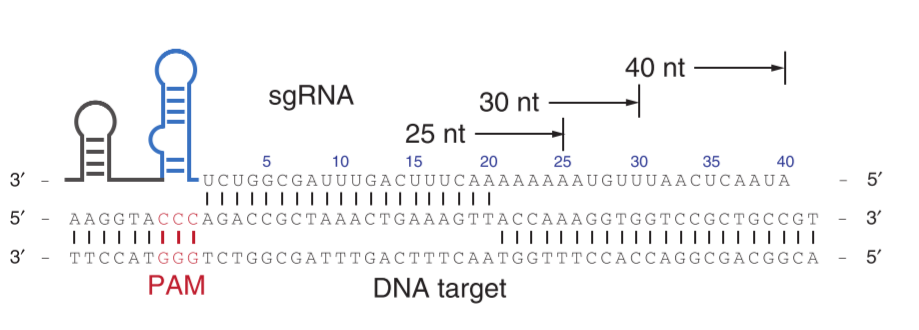
Input or parameter chosen:
- Gene cds sequence: Human EPO cds (GenBank: BC143225.1)
- Type of Cas: dxCas9
- PAM sequence: NG
- sgRNA length: 20 bp of target sgRNA
- Off target possibility (seed to target): 10 bp (50 % adjacent to - PAM should be identical)
Process:
1. Translate from target DNA to protein.
2. Calculate the number of possible codons for each amio acid.
3. Expand the initial tartget DNA to a set of DNA, with various codon possibilities. (product of the numebr of possibility in the sequence)
4. Record the all possible position of the PAM site within the 20 bp distance forward the exon-exon junction.
5. Determine the total number of sgRNAs necessary for each PAM site position.
6. Select the PAM sequence position that has the minimal number of sgRNAs possible and generate sgRNAs.
Cons:
MATLAB ???
Gene Doping Model
- (Optional) From muscle to blood.
- From blood to tissue.
- Take degradation and diffusion into considration.
- Process of Virus into the cell and the gene delivery.
- The expression of the EPO gene.
- The process of blood cell formation.
Human Practice
- We implemented athletes' wishes regarding invasivity, privacy and testing frequency into our detection method.
- Further interaction with stakeholders such as the Dutch Doping Authority and Oxford Nanopore Technologies made us add a **prescreen and a barcoding tool to our detection method for versatile, high throughput and reliable detection.
- We invited engineers to hack our detection method, and used their collective strength to improve our algorithm and anticipate future gene doping developments.

Overall:



