@HaomingJiang
2018-03-01T04:12:26.000000Z
字数 23568
阅读 1569
HW 3
Haoming Jiang
Ex 3.1
a
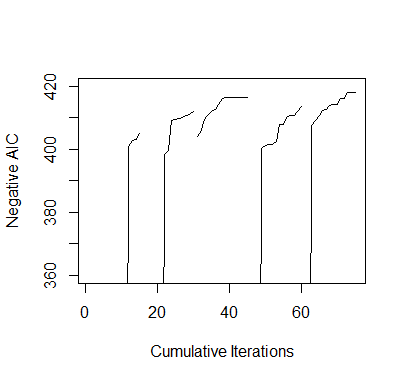
> #########################################################################> ### EXERCISE 3.1> #########################################################################> # baseball.dat = entire data set> # baseball.sub = matrix of all predictors> # salary.log = response, log salary> # n = number of observations in the data set> # m = number of predictors in the data set> # num.starts = number of random starts> # runs = matrix containing the random starts where each row is a> # vector of the parameters included for the model> # (1 = included, 0 = omitted)> # runs.aic = AIC values for the best model found for each> # of the random starts> # itr = number of iterations for each random start> #########################################################################>> ## INITIAL VALUES> baseball.dat = read.table("baseball.dat",header=TRUE)> baseball.dat$freeagent = factor(baseball.dat$freeagent)> baseball.dat$arbitration = factor(baseball.dat$arbitration)> baseball.sub = baseball.dat[,-1]> salary.log = log(baseball.dat$salary)> n = length(salary.log)> m = length(baseball.sub[1,])> num.starts = 5> runs = matrix(0,num.starts,m)> itr = 15> runs.aic = matrix(0,num.starts,itr)>> # INITIALIZES STARTING RUNS> set.seed(19676)> for(i in 1:num.starts){runs[i,] = rbinom(m,1,.5)}>> ## MAIN> for(k in 1:num.starts){+ run.current = runs[k,]++ # ITERATES EACH RANDOM START+ for(j in 1:itr){+ run.vars = baseball.sub[,run.current==1]+ g = lm(salary.log~.,run.vars)+ run.aic = extractAIC(g)[2]+ run.next = run.current++ # TESTS ALL MODELS IN THE 1-NEIGHBORHOOD AND SELECTS THE+ # MODEL WITH THE LOWEST AIC+ for(i in sample(m)){+ run.step = run.current+ run.step[i] = !run.current[i]+ run.vars = baseball.sub[,run.step==1]+ g = lm(salary.log~.,run.vars)+ run.step.aic = extractAIC(g)[2]+ if(run.step.aic < run.aic){+ run.next = run.step+ run.aic = run.step.aic+ break+ }+ }+ run.current = run.next+ runs.aic[k,j]=run.aic+ }+ runs[k,] = run.current+ }>> ## OUTPUT> runs # LISTS OF PREDICTORS[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14] [,15] [,16][1,] 1 0 0 1 0 0 1 0 1 1 0 0 1 1 1 0[2,] 1 0 1 0 0 1 0 1 0 1 1 0 1 1 1 1[3,] 0 0 1 0 0 1 0 1 0 1 0 0 1 1 1 0[4,] 1 1 1 0 0 1 0 1 0 1 0 0 1 1 1 1[5,] 1 0 1 0 0 0 0 1 0 1 0 0 1 1 1 1[,17] [,18] [,19] [,20] [,21] [,22] [,23] [,24] [,25] [,26] [,27][1,] 0 1 1 0 0 0 0 0 1 0 1[2,] 1 1 0 0 1 0 0 1 0 1 1[3,] 0 0 0 1 1 1 0 0 1 1 0[4,] 0 0 0 0 0 1 1 1 1 1 0[5,] 0 0 0 0 0 0 0 1 1 0 1> runs.aic # AIC VALUES[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8][1,] -217.9065 -217.9935 -219.8484 -220.1619 -221.0135 -222.6077 -223.7776 -223.9698[2,] -217.2085 -218.3258 -219.5092 -219.7432 -221.7374 -223.6676 -398.4278 -399.6693[3,] -403.9695 -405.3088 -409.3507 -410.7968 -411.9632 -412.6045 -414.2432 -416.2141[4,] -124.5394 -127.0578 -127.5083 -400.2682 -401.1612 -401.6403 -401.6661 -402.5395[5,] -135.9155 -137.7239 -407.5777 -409.1986 -410.8303 -412.4298 -412.6726 -413.9250[,9] [,10] [,11] [,12] [,13] [,14] [,15][1,] -225.8663 -232.0466 -233.5325 -400.9725 -402.8854 -403.2838 -404.8729[2,] -409.1566 -409.3218 -409.6924 -410.1146 -410.6694 -411.0543 -412.0218[3,] -416.5069 -416.5069 -416.5069 -416.5069 -416.5069 -416.5069 -416.5069[4,] -407.9254 -407.9608 -410.2451 -410.6919 -410.8825 -411.9445 -413.5147[5,] -414.0846 -414.2130 -416.1533 -416.2576 -417.8858 -417.8858 -417.8858>> ##PLOT> plot(1:(itr*num.starts),-c(t(runs.aic)),xlab="Cumulative Iterations",+ ylab="Negative AIC",ylim=c(360,420),type="n")> for(i in 1:num.starts) {+ lines((i-1)*itr+(1:itr),-runs.aic[i,]) }
b
It takes longer time. And AIC are much greater than the previouse ones. All because somtimes it can not find a downhill 2-neighborhood.
> #########################################################################> ### EXERCISE 3.1> #########################################################################> # baseball.dat = entire data set> # baseball.sub = matrix of all predictors> # salary.log = response, log salary> # n = number of observations in the data set> # m = number of predictors in the data set> # num.starts = number of random starts> # runs = matrix containing the random starts where each row is a> # vector of the parameters included for the model> # (1 = included, 0 = omitted)> # runs.aic = AIC values for the best model found for each> # of the random starts> # itr = number of iterations for each random start> #########################################################################>> ## INITIAL VALUES> baseball.dat = read.table("baseball.dat",header=TRUE)> baseball.dat$freeagent = factor(baseball.dat$freeagent)> baseball.dat$arbitration = factor(baseball.dat$arbitration)> baseball.sub = baseball.dat[,-1]> salary.log = log(baseball.dat$salary)> n = length(salary.log)> m = length(baseball.sub[1,])> num.starts = 5> runs = matrix(0,num.starts,m)> itr = 15> runs.aic = matrix(0,num.starts,itr)>> # INITIALIZES STARTING RUNS> set.seed(19676)> for(i in 1:num.starts){runs[i,] = rbinom(m,1,.5)}>> all2combn = combn(m,2)>> ## MAIN> for(k in 1:num.starts){+ run.current = runs[k,]++ # ITERATES EACH RANDOM START+ for(j in 1:itr){+ run.vars = baseball.sub[,run.current==1]+ g = lm(salary.log~.,run.vars)+ run.aic = extractAIC(g)[2]+ run.next = run.current++ # TESTS ALL MODELS IN THE 1-NEIGHBORHOOD AND SELECTS THE+ # MODEL WITH THE LOWEST AIC+ for(i in sample(m*(m-1)/2)){+ run.step = run.current+ run.step[all2combn[1][i]] = !run.current[all2combn[1][i]]+ run.step[all2combn[2][i]] = !run.current[all2combn[2][i]]+ run.vars = baseball.sub[,run.step==1]+ g = lm(salary.log~.,run.vars)+ run.step.aic = extractAIC(g)[2]+ if(run.step.aic < run.aic){+ run.next = run.step+ run.aic = run.step.aic+ break+ }+ }+ run.current = run.next+ runs.aic[k,j]=run.aic+ }+ runs[k,] = run.current+ }>> ## OUTPUT> runs # LISTS OF PREDICTORS[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14] [,15] [,16][1,] 1 0 0 1 1 1 0 0 0 0 0 1 1 0 0 0[2,] 1 1 0 0 0 1 1 1 1 1 1 0 1 0 0 1[3,] 1 0 0 0 1 0 0 1 0 1 1 0 1 1 1 1[4,] 1 0 1 0 0 1 1 0 1 1 0 0 0 1 0 1[5,] 1 1 1 1 1 0 1 1 1 1 0 1 0 1 1 0[,17] [,18] [,19] [,20] [,21] [,22] [,23] [,24] [,25] [,26] [,27][1,] 0 1 1 0 1 1 0 0 1 0 0[2,] 1 1 0 1 0 0 0 1 1 1 1[3,] 0 0 0 0 0 1 0 0 1 0 0[4,] 0 1 1 0 1 1 1 1 0 0 0[5,] 0 0 0 1 0 1 1 1 1 0 1> runs.aic # AIC VALUES[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8][1,] -217.2883 -217.2883 -217.2883 -217.2883 -217.2883 -217.2883 -217.2883 -217.2883[2,] -216.9405 -216.9405 -216.9405 -216.9405 -216.9405 -216.9405 -216.9405 -216.9405[3,] -400.3062 -400.3062 -400.3062 -400.3062 -400.3062 -400.3062 -400.3062 -400.3062[4,] -122.8772 -122.8772 -122.8772 -122.8772 -122.8772 -122.8772 -122.8772 -122.8772[5,] -136.5481 -136.5481 -136.5481 -136.5481 -136.5481 -136.5481 -136.5481 -136.5481[,9] [,10] [,11] [,12] [,13] [,14] [,15][1,] -217.2883 -217.2883 -217.2883 -217.2883 -217.2883 -217.2883 -217.2883[2,] -216.9405 -216.9405 -216.9405 -216.9405 -216.9405 -216.9405 -216.9405[3,] -400.3062 -400.3062 -400.3062 -400.3062 -400.3062 -400.3062 -400.3062[4,] -122.8772 -122.8772 -122.8772 -122.8772 -122.8772 -122.8772 -122.8772[5,] -136.5481 -136.5481 -136.5481 -136.5481 -136.5481 -136.5481 -136.5481>> ##PLOT> plot(1:(itr*num.starts),-c(t(runs.aic)),xlab="Cumulative Iterations",+ ylab="Negative AIC",ylim=c(360,420),type="n")> for(i in 1:num.starts) {+ lines((i-1)*itr+(1:itr),-runs.aic[i,]) }
Ex 3.8
We can just do kmeans, which can find a local minima.
The size of clusters are 67,62,49
The following is the visualization of the clustering in 2D space via PCA.
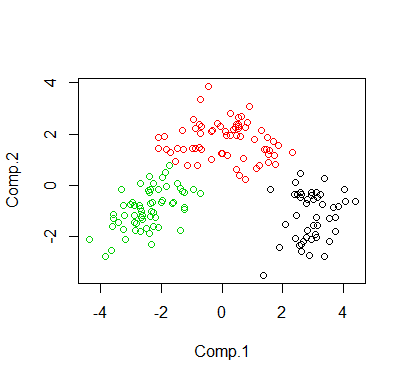
> wine.dat = read.table("wine.dat",header=TRUE)> cl <- kmeans(wine.dat, 3)> print(cl$size)[1] 49 67 62>> # plot in 2D via PCA> pcafit <- princomp(wine.dat)> plot(pcafit$scores[,1:2], col = cl$cluster)
Ex 4.1
a&b
The M-step is the same. But for E-step, we use and to esitmate the expexted genotype frequencies instead and .
Result:
# FINAL ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)[1] 0.03606708 0.19579915 0.76813377
> #########################################################################> # x = observed phenotype counts (carbonaria, insularia, typica, i or t)> # n = expected genotype frequencies (CC, CI, CT, II, IT, TT)> # p = allele probabilities (carbonaria, insularia, typica)> # itr = number of iterations> # allele.e = computes expected genotype frequencies> # allele.m = computes allele probabilities> #########################################################################>> ## INITIAL VALUES> x = c(85, 196, 341, 578)> n = rep(0,6)> p = rep(1/3,3)> itr = 40>> ## EXPECTATION AND MAXIMIZATION FUNCTIONS> allele.e = function(x,p){+ n.cc = (x[1]*(p[1]^2))/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ n.ci = (2*x[1]*p[1]*p[2])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ n.ct = (2*x[1]*p[1]*p[3])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ x2hat = x[2]+x[4]*((p[2]^2)+2*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])+ x3hat = x[3]+x[4]*(p[3]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])+ n.ii = (x2hat*(p[2]^2))/((p[2]^2)+2*p[2]*p[3])+ n.it = (2*x2hat*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3])+ n = c(n.cc,n.ci,n.ct,n.ii,n.it,x3hat)+ return(n)+ }>> allele.m = function(x,n){+ p.c = (2*n[1]+n[2]+n[3])/(2*sum(x))+ p.i = (2*n[4]+n[5]+n[2])/(2*sum(x))+ p.t = (2*n[6]+n[3]+n[5])/(2*sum(x))+ p = c(p.c,p.i,p.t)+ return(p)+ }>> ## MAIN> for(i in 1:itr){+ n = allele.e(x,p)+ p = allele.m(x,n)+ }>> ## OUTPUT> p # FINAL ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)[1] 0.03606708 0.19579915 0.76813377
c
Result:
> ## OUTPUT> sd.hat # STANDARD ERROR ESTIMATES (pc, pi, pt)[1] 0.003840308 0.011077418 0.011530716> cor.hat # CORRELATION ESTIMATES ({pc,pi}, {pc,pt}, {pi,pt})[1] -0.03341484 -0.30094901 -0.94955901
Code:
> #########################################################################> # x = observed phenotype counts (carbonaria, insularia, typica)> # n = expected genotype frequencies (CC, CI, CT, II, IT, TT)> # p = allele probabilities (carbonaria, insularia, typica)> # p.em = em algorithm estimates> # n.em = em algorithm estimates> # itr = number of iterations> # allele.e = computes expected genotype frequencies> # allele.m = computes allele probabilities> #########################################################################>> ## INITIAL VALUES> x = c(85, 196, 341, 578)> n = rep(0,6)> itr = 40> p = c(0.04, 0.19, 0.77)> p.em = p> theta = matrix(0,3,3)> psi = rep(0,3)> r = matrix(0,3,3)>> ## EXPECTATION AND MAXIMIZATION FUNCTIONS> allele.e = function(x,p){+ n.cc = (x[1]*(p[1]^2))/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ n.ci = (2*x[1]*p[1]*p[2])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ n.ct = (2*x[1]*p[1]*p[3])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ x2hat = x[2]+x[4]*((p[2]^2)+2*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])+ x3hat = x[3]+x[4]*(p[3]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])+ n.ii = (x2hat*(p[2]^2))/((p[2]^2)+2*p[2]*p[3])+ n.it = (2*x2hat*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3])+ n = c(n.cc,n.ci,n.ct,n.ii,n.it,x3hat)+ return(n)+ }>> allele.m = function(x,n){+ p.c = (2*n[1]+n[2]+n[3])/(2*sum(x))+ p.i = (2*n[4]+n[5]+n[2])/(2*sum(x))+ p.t = (2*n[6]+n[3]+n[5])/(2*sum(x))+ p = c(p.c,p.i,p.t)+ return(p)+ }>> ## COMPUTES EM ALGORITHM ESTIMATES> for(i in 1:itr){+ n.em = allele.e(x,p.em)+ p.em = allele.m(x,n.em)+ }>> ## INTIALIZES THETA> for(j in 1:length(p)){+ theta[,j] = p.em+ theta[j,j] = p[j]+ }>> ## MAIN> for(t in 1:5){+ n = allele.e(x,p)+ p.hat = allele.m(x,n)+ for(j in 1:length(p)){+ theta[j,j] = p.hat[j]+ n = allele.e(x,theta[,j])+ psi = allele.m(x,n)+ for(i in 1:length(p)){+ r[i,j] = (psi[i]-p.em[i])/(theta[j,j]-p.em[j])+ }+ }+ p = p.hat+ }>> ## COMPLETE INFORMATION> iy.hat=matrix(0,2,2)> iy.hat[1,1] = ((2*n.em[1]+n.em[2]+n.em[3])/(p.em[1]^2) ++ (2*n.em[6]+n.em[3]+n.em[5])/(p.em[3]^2))> iy.hat[2,2] = ((2*n.em[4]+n.em[5]+n.em[2])/(p.em[2]^2) ++ (2*n.em[6]+n.em[3]+n.em[5])/(p.em[3]^2))> iy.hat[1,2] = iy.hat[2,1] = (2*n.em[6]+n.em[3]+n.em[5])/(p.em[3]^2)>> ## COMPUTES STANDARD ERRORS AND CORRELATIONS> var.hat = solve(iy.hat)%*%(diag(2)+t(r[-3,-3])%*%solve(diag(2)-t(r[-3,-3])))> sd.hat = c(sqrt(var.hat[1,1]),sqrt(var.hat[2,2]),sqrt(sum(var.hat)))> cor.hat = c(var.hat[1,2]/(sd.hat[1]*sd.hat[2]),+ (-var.hat[1,1]-var.hat[1,2])/(sd.hat[1]*sd.hat[3]),+ (-var.hat[2,2]-var.hat[1,2])/(sd.hat[2]*sd.hat[3]))>> ## OUTPUT> sd.hat # STANDARD ERROR ESTIMATES (pc, pi, pt)[1] 0.003840308 0.011077418 0.011530716> cor.hat # CORRELATION ESTIMATES ({pc,pi}, {pc,pt}, {pi,pt})[1] -0.03341484 -0.30094901 -0.94955901
d
Result:
> ## OUTPUT> theta[,1] # EM ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)[1] 0.03606708 0.19579915 0.76813377> sd.hat # STANDARD ERROR ESTIMATES (p.c, p.i, p.t)[1] 0.003864236 0.012547943 0.012905975> cor.hat # CORRELATION ESTIMATES ({p.c,p.i}, {p.c,p.t}, {p.i,p.t})[1] -0.06000425 -0.24107484 -0.95429228
Code:
> #########################################################################> # x = observed phenotype counts (carbonaria, insularia, typica)> # n = expected genotype frequencies (CC, CI, CT, II, IT, TT)> # p = allele probabilities (carbonaria, insularia, typica)> # itr = number of iterations> # theta = allele probabilities for psuedo-data> # allele.e = computes expected genotype frequencies> # allele.m = computes allele probabilities> #########################################################################>> ## INITIAL VALUES> x = c(85, 196, 341,578)> n = rep(0,6)> p = rep(1/3,3)> itr = 40> theta = matrix(0,3,10000)> set.seed(0)>> ## EXPECTATION AND MAXIMIZATION FUNCTIONS> allele.e = function(x,p){+ n.cc = (x[1]*(p[1]^2))/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ n.ci = (2*x[1]*p[1]*p[2])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ n.ct = (2*x[1]*p[1]*p[3])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])+ x2hat = x[2]+x[4]*((p[2]^2)+2*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])+ x3hat = x[3]+x[4]*(p[3]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])+ n.ii = (x2hat*(p[2]^2))/((p[2]^2)+2*p[2]*p[3])+ n.it = (2*x2hat*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3])+ n = c(n.cc,n.ci,n.ct,n.ii,n.it,x3hat)+ return(n)+ }> allele.m = function(x,n){+ p.c = (2*n[1]+n[2]+n[3])/(2*sum(x))+ p.i = (2*n[4]+n[5]+n[2])/(2*sum(x))+ p.t = (2*n[6]+n[3]+n[5])/(2*sum(x))+ p = c(p.c,p.i,p.t)+ return(p)+ }>> ## MAIN> for(i in 1:itr){+ n = allele.e(x,p)+ p = allele.m(x,n)+ }> theta[,1] = p> for(j in 2:10000){+ n.c = rbinom(1, sum(x), x[1]/sum(x))+ n.i = rbinom(1, sum(x) - n.c, x[2]/(sum(x)-x[1]))+ n.t = rbinom(1, sum(x) - n.c - n.i, x[3]/(sum(x)-x[1]-x[2]))+ n.u = sum(x) - n.c - n.i - n.t+ x.new = c(n.c, n.i, n.t, n.u)+ n = rep(0,6)+ p = rep(1/3,3)+ for(i in 1:itr){+ n = allele.e(x.new,p)+ p = allele.m(x.new,n)+ }+ theta[,j] = p+ }>> sd.hat = c(sd(theta[1,]), sd(theta[2,]), sd(theta[3,]))> cor.hat = c(cor(theta[1,],theta[2,]), cor(theta[1,],theta[3,]),+ cor(theta[2,],theta[3,]))>> ## OUTPUT> theta[,1] # EM ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)[1] 0.03606708 0.19579915 0.76813377> sd.hat # STANDARD ERROR ESTIMATES (p.c, p.i, p.t)[1] 0.003864236 0.012547943 0.012905975> cor.hat # CORRELATION ESTIMATES ({p.c,p.i}, {p.c,p.t}, {p.i,p.t})[1] -0.06000425 -0.24107484 -0.95429228
e
Output:
> ## OUTPUT> p # FINAL ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)[1] 0.03596147 0.19582060 0.76821793
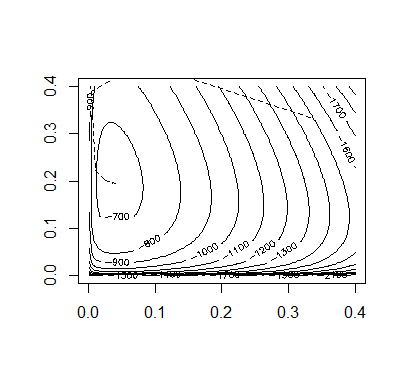
Code:
## INITIAL VALUESx = c(85, 196, 341, 578)n = rep(0,6)p = rep(1/3,3)itr = 40prob.values = matrix(0,3,itr+1)prob.values[,1] = palpha.default = 2## FUNCTIONSallele.e = function(x,p){n.cc = (x[1]*(p[1]^2))/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])n.ci = (2*x[1]*p[1]*p[2])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])n.ct = (2*x[1]*p[1]*p[3])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])x2hat = x[2]+x[4]*((p[2]^2)+2*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])x3hat = x[3]+x[4]*(p[3]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])n.ii = (x2hat*(p[2]^2))/((p[2]^2)+2*p[2]*p[3])n.it = (2*x2hat*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3])n = c(n.cc,n.ci,n.ct,n.ii,n.it,x3hat)return(n)}allele.l = function(x,p){l = ( x[1]*log(2*p[1] - p[1]^2) + x[2]*log(p[2]^2 + 2*p[2]*p[3]) +2*x[3]*log(p[3]) + x[4]*log(p[2]^2 + 2*p[2]*p[3]+p[3]^2) )return(l)}Q.prime = function(n,p){da = (2*n[1]+n[2]+n[3])/(p[1]) - (2*n[6]+n[3]+n[5])/(p[3])db = (2*n[4]+n[5]+n[2])/(p[2]) - (2*n[6]+n[3]+n[5])/(p[3])dQ = c(da,db)return(dQ)}Q.2prime = function(n,p){da2 = -(2*n[1]+n[2]+n[3])/(p[1]^2) - (2*n[6]+n[3]+n[5])/(p[3]^2)51db2 = -(2*n[4]+n[5]+n[2])/(p[2]^2) - (2*n[6]+n[3]+n[5])/(p[3]^2)dab = -(2*n[6]+n[3]+n[5])/(p[3]^2)d2Q = matrix(c(da2,dab,dab,db2), nrow=2, byrow=TRUE)return(d2Q)}## MAINl.old = allele.l(x,p)for(i in 1:itr){alpha = alpha.defaultn = allele.e(x,p)p.new = p[1:2] - alpha*solve(Q.2prime(n,p))%*%Q.prime(n,p)p.new[3] = 1 - p.new[1] - p.new[2]if(p.new > 0 && p.new < 1){l.new = allele.l(x,p.new)}# REDUCE ALPHA UNTIL A CORRECT STEP IS REACHEDwhile(p.new < 0 || p.new > 1 || l.new < l.old){alpha = alpha/2p.new = p[1:2] - alpha*solve(Q.2prime(n,p))%*%Q.prime(n,p)p.new[3] = 1 - p.new[1] - p.new[2]if(p.new > 0 && p.new < 1){l.new = allele.l(x,p.new)}}p = p.newprob.values[,i+1] = pl.old = l.new}## OUTPUTp # FINAL ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)## PLOT OF CONVERGENCEpb = seq(0.001,0.4,length=100)c = outer(pb,pb,function(a,b){1-a-b})pbs = matrix(0,3,10000)pbs[1,] = rep(pb,100)pbs[2,] = rep(pb,each=100)pbs[3,] = as.vector(c)z = matrix(apply(pbs,2,allele.l,x=x),100,100)contour(pb,pb,z,nlevels=20)for(i in 1:itr){segments(prob.values[1,i],prob.values[2,i],prob.values[1,i+1],prob.values[2,i+1],lty=2)}
f
Output:
> ## OUTPUT> p # FINAL ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)[1] 0.03606708 0.19579915 0.76813377
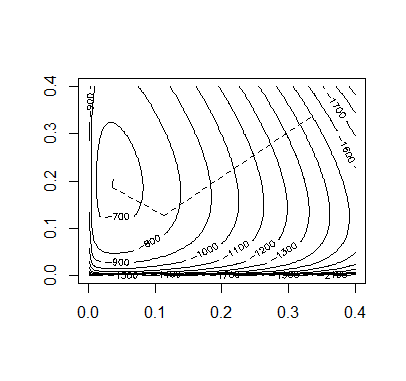
Code:
## INITIAL VALUESx = c(85, 196, 341,578)n = rep(0,6)p = rep(1/3,3)itr = 40prob.values = matrix(0,3,itr+1)prob.values[,1] = palpha.default = 2## FUNCTIONSallele.e = function(x,p){n.cc = (x[1]*(p[1]^2))/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])n.ci = (2*x[1]*p[1]*p[2])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])n.ct = (2*x[1]*p[1]*p[3])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])x2hat = x[2]+x[4]*((p[2]^2)+2*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])x3hat = x[3]+x[4]*(p[3]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])n.ii = (x2hat*(p[2]^2))/((p[2]^2)+2*p[2]*p[3])n.it = (2*x2hat*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3])n = c(n.cc,n.ci,n.ct,n.ii,n.it,x3hat)return(n)}allele.m = function(x,n){p.c = (2*n[1]+n[2]+n[3])/(2*sum(x))p.i = (2*n[4]+n[5]+n[2])/(2*sum(x))p.t = (2*n[6]+n[3]+n[5])/(2*sum(x))p = c(p.c,p.i,p.t)return(p)}allele.l = function(x,p){l = ( x[1]*log(2*p[1] - p[1]^2) + x[2]*log(p[2]^2 + 2*p[2]*p[3]) +2*x[3]*log(p[3]) + 2*x[4]*log(1-p[1]) )return(l)}allele.iy = function(n,p){iy.hat=matrix(0,2,2)iy.hat[1,1] = ((2*n[1]+n[2]+n[3])/(p[1]^2) +(2*n[6]+n[3]+n[5])/(p[3]^2))iy.hat[2,2] = ((2*n[4]+n[5]+n[2])/(p[2]^2) +(2*n[6]+n[3]+n[5])/(p[3]^2))iy.hat[1,2] = iy.hat[2,1] = (2*n[6]+n[3]+n[5])/(p[3]^2)return(iy.hat)}allele.l.2prime = function(x,p){l.2prime = matrix(0,2,2)l.2prime[1,1] = ( (-x[1]*(2-2*p[1])^2)/((2*p[1]-p[1]^2)^2) -2*x[1]/(2*p[1]-p[1]^2) -(4*x[2])/((-2*p[1]-p[2]+2)^2) -2*x[3]/(p[3]^2)-2*x[4]/(1-p[1])^2)l.2prime[2,2] = ( (-4*x[2]*p[3]^2)/((p[2]^2 + 2*p[2]*p[3])^2) -2*x[2]/(p[2]^2 + 2*p[2]*p[3]) -2*x[3]/(p[3]^2))l.2prime[1,2] = ((-2*x[2])/((-2*p[1]-p[2]+2)^2) -2*x[3]/(p[3]^2))l.2prime[2,1] = l.2prime[1,2]return(l.2prime)}## MAINl.old = allele.l(x,p)for(i in 1:itr){alpha = alpha.defaultn = allele.e(x,p)p.em = allele.m(x,n)p.new = (p[1:2] - alpha*solve(allele.l.2prime(x,p))%*%allele.iy(n,p)%*%(p.em[1:2]-p[1:2]))p.new[3] = 1 - p.new[1] - p.new[2]if(p.new > 0 && p.new < 1){l.new = allele.l(x,p.new)}# REDUCE ALPHA UNTIL A CORRECT STEP IS REACHEDwhile(p.new < 0 || p.new > 1 || l.new < l.old){alpha = alpha/2p.new = (p[1:2] - alpha*solve(allele.l.2prime(x,p))%*%allele.iy(n,p)%*%(p.em[1:2]-p[1:2]))p.new[3] = 1 - p.new[1] - p.new[2]if(p.new > 0 && p.new < 1){l.new = allele.l(x,p.new)}}p = p.newprob.values[,i+1] = pl.old = l.new}## OUTPUTp # FINAL ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)## PLOT OF CONVERGENCEpb = seq(0.001,0.4,length=100)c = outer(pb,pb,function(a,b){1-a-b})pbs = matrix(0,3,10000)pbs[1,] = rep(pb,100)pbs[2,] = rep(pb,each=100)pbs[3,] = as.vector(c)z = matrix(apply(pbs,2,allele.l,x=x),100,100)contour(pb,pb,z,nlevels=20)for(i in 1:itr){segments(prob.values[1,i],prob.values[2,i],prob.values[1,i+1],prob.values[2,i+1],lty=2)}
g
Output:
> ## OUTPUT> p # FINAL ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)[1] 0.03606708 0.19580543 0.76812749
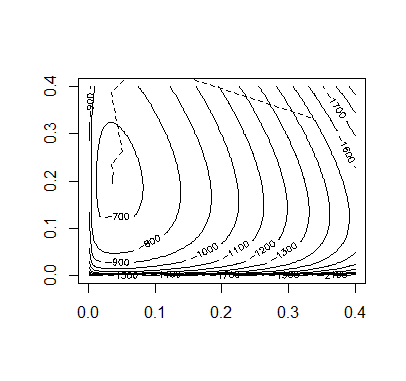
Code:
## INITIAL VALUESx = c(85, 196, 341, 578)n = rep(0,6)p = rep(1/3,3)itr = 20m = matrix(0,2,2)b = matrix(0,2,2)prob.values = matrix(0,3,itr+1)prob.values[,1] = palpha.default = 2## FUNCTIONSallele.e = function(x,p){n.cc = (x[1]*(p[1]^2))/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])n.ci = (2*x[1]*p[1]*p[2])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])n.ct = (2*x[1]*p[1]*p[3])/((p[1]^2)+2*p[1]*p[2]+2*p[1]*p[3])x2hat = x[2]+x[4]*((p[2]^2)+2*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])x3hat = x[3]+x[4]*(p[3]*p[3])/((p[2]^2)+2*p[2]*p[3]+p[3]*p[3])n.ii = (x2hat*(p[2]^2))/((p[2]^2)+2*p[2]*p[3])n.it = (2*x2hat*p[2]*p[3])/((p[2]^2)+2*p[2]*p[3])n = c(n.cc,n.ci,n.ct,n.ii,n.it,x3hat)return(n)}allele.l = function(x,p){l = ( x[1]*log(2*p[1] - p[1]^2) + x[2]*log(p[2]^2 + 2*p[2]*p[3]) +2*x[3]*log(p[3]) + 2*x[4]*log(1-p[1]) )return(l)}Q.prime = function(n,p){da = (2*n[1]+n[2]+n[3])/(p[1]) - (2*n[6]+n[3]+n[5])/(p[3])db = (2*n[4]+n[5]+n[2])/(p[2]) - (2*n[6]+n[3]+n[5])/(p[3])dQ = c(da,db)return(dQ)}Q.2prime = function(n,p){da2 = -(2*n[1]+n[2]+n[3])/(p[1]^2) - (2*n[6]+n[3]+n[5])/(p[3]^2)db2 = -(2*n[4]+n[5]+n[2])/(p[2]^2) - (2*n[6]+n[3]+n[5])/(p[3]^2)dab = -(2*n[6]+n[3]+n[5])/(p[3]^2)d2Q = matrix(c(da2,dab,dab,db2), nrow=2, byrow=TRUE)return(d2Q)}## MAINl.old = allele.l(x,p)for(i in 1:itr){alpha = alpha.defaultn = allele.e(x,p)m = Q.2prime(n,p) - bp.new = p[1:2] - alpha*solve(m)%*%Q.prime(n,p)p.new[3] = 1 - p.new[1] - p.new[2]if(p.new > 0 && p.new < 1){l.new = allele.l(x,p.new)}# REDUCE ALPHA UNTIL A CORRECT STEP IS REACHEDwhile(p.new < 0 || p.new > 1 || l.new < l.old){alpha = alpha/2p.new = p[1:2] - alpha*solve(m)%*%Q.prime(n,p)p.new[3] = 1 - p.new[1] - p.new[2]if(p.new > 0 && p.new < 1){l.new = allele.l(x,p.new)}}at = p.new[1:2]-p[1:2]n = allele.e(x,p.new)bt = Q.prime(n,p)-Q.prime(n,p.new)vt = bt - b%*%atct = as.numeric(1/(t(vt)%*%at))b = b + ct*vt%*%t(vt)p = p.newprob.values[,i+1] = pl.old = l.new}## OUTPUTp # FINAL ESTIMATE FOR ALLELE PROBABILITIES (p.c, p.i, p.t)## PLOT OF CONVERGENCEpb = seq(0.001,0.4,length=100)c = outer(pb,pb,function(a,b){1-a-b})pbs = matrix(0,3,10000)pbs[1,] = rep(pb,100)pbs[2,] = rep(pb,each=100)pbs[3,] = as.vector(c)z = matrix(apply(pbs,2,allele.l,x=x),100,100)contour(pb,pb,z,nlevels=20)for(i in 1:itr){segments(prob.values[1,i],prob.values[2,i],prob.values[1,i+1],prob.values[2,i+1],lty=2)}
h
Accoding to the figures of the above methods, we can tell the Aitken accerlerated EM is the most efficient.
4.5
a
Forward function:
Given for all . Then for any , we compute
Backward function:
Given for all . Then for any , we compute
b
C
The log-likelihood is :
So we can take and as laten variables. The proposed fomular is the according MLE when fixing and .
d
Output:
$pi[1] 0.5 0.5$p[,1] [,2][1,] 0.5 0.5[2,] 0.5 0.5$e[,1] [,2][1,] 0.45 0.55[2,] 0.45 0.55
Code
x = as.matrix(read.table("coin.dat",header=TRUE))+1n = 200pi = c(0.5,0.5)e = matrix(rep(0.5,4),ncol=2)p = matrix(rep(0.5,4),ncol=2)cal_alpha = function(pi,e,p,x){alpha_new = matrix(rep(0,n*2),ncol=2)alpha_new[1,1] = pi[1]*e[1,x[1]]alpha_new[1,2] = pi[2]*e[2,x[1]]for (i in 2:n){alpha_new[i,1] = alpha_new[i-1,1]*p[1,1]*e[1,x[i]] + alpha_new[i-1,2]*p[2,1]*e[1,x[i]]alpha_new[i,2] = alpha_new[i-1,1]*p[1,2]*e[2,x[i]] + alpha_new[i-1,2]*p[2,2]*e[2,x[i]]}return(alpha_new)}cal_beta = function(pi,e,p,x){beta_new = matrix(rep(0,n*2),ncol=2)beta_new[n,1] = 1beta_new[n,2] = 1for (i in (n-1):1){beta_new[i,1] = p[1,1]*e[1,x[i+1]]*beta_new[i+1,1] + p[1,2]*e[2,x[i+1]]*beta_new[i+1,2]beta_new[i,2] = p[2,1]*e[1,x[i+1]]*beta_new[i+1,1] + p[2,2]*e[2,x[i+1]]*beta_new[i+1,2]}return(beta_new)}estep = function(x,alpha,beta,p,e){Nh = c(0,0)Nhh = matrix(c(0,0,0,0),ncol=2)Nho = matrix(c(0,0,0,0),ncol=2)Nh = c(alpha[1,1]*beta[1,1],alpha[1,2]*beta[1,2])for(i in 1:2)for(j in 1:2)for(ii in 1:(n-1))Nhh[i,j] = Nhh[i,j]+alpha[ii,i]*p[i,j]*e[j,x[ii+1]]*beta[ii+1,j]for(i in 1:2)for(j in 1:2){for(ii in 1:(n))if(x[ii] == j){Nho[i,j] = Nho[i,j]+alpha[ii,i]*beta[ii,i]}}return(list(Nh=Nh,Nhh=Nhh,Nho=Nho))}mstep = function(Nh,Nhh,Nho){new_pi = Nh/sum(Nh)new_p = Nhh/rowSums(Nhh)new_e = Nho/rowSums(Nho)return(list(pi=new_pi,p=new_p,e=new_e))}iternum = 10for(iter in 1:iternum){alpha = cal_alpha(pi,e,p,x)beta = cal_beta(pi,e,p,x)estep_list = estep(x,alpha,beta,p,e)mstep_list = mstep(estep_list$Nh,estep_list$Nhh,estep_list$Nho)pi = mstep_list$pip = mstep_list$pe = mstep_list$eprint(mstep_list)}
